Method for identifying predator nematophagous hyphomycete arthrobotrys through DNA bar codes
A technique for Arthrospora and Mycelia, which is applied in the field of fungal species identification, and achieves the effects of low completeness requirements, easy comparison, and improved identification efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment 1: The establishment of the gene bank of the RPB2 gene sequence fragment that preys on the nematode hyphomycetes Arthrobush sp. DNA barcode
[0019] (1) There are 32 species and 54 strains of the selected strains that prey on the nematode hyphomycete Arthrobush, all of which are recorded in "Arthroxa and related genera" in Volume 33 of "Mycology of China" Species with a clear concept of spores, and each common species contains at least 3 strains to meet the needs of the subsequent calculation of the maximum genetic variation value within the species. See Table 1 for details.
[0020] Table 1 Sample information sheet
[0021]
[0022]
[0023]
[0024] (2) Inoculate the pure culture strains of the samples listed in Table 1 on a CMA plate [corn flour medium: 30 g of corn flour, 16 g of agar, 1000 ml of water, natural pH, corn flour and an appropriate amount of water and boil for 30 minutes, filter with four layers of gauze, and the filtrate Add water...
Embodiment 2
[0028] Example 2: Identification of unknown species of Arthrobush fungus
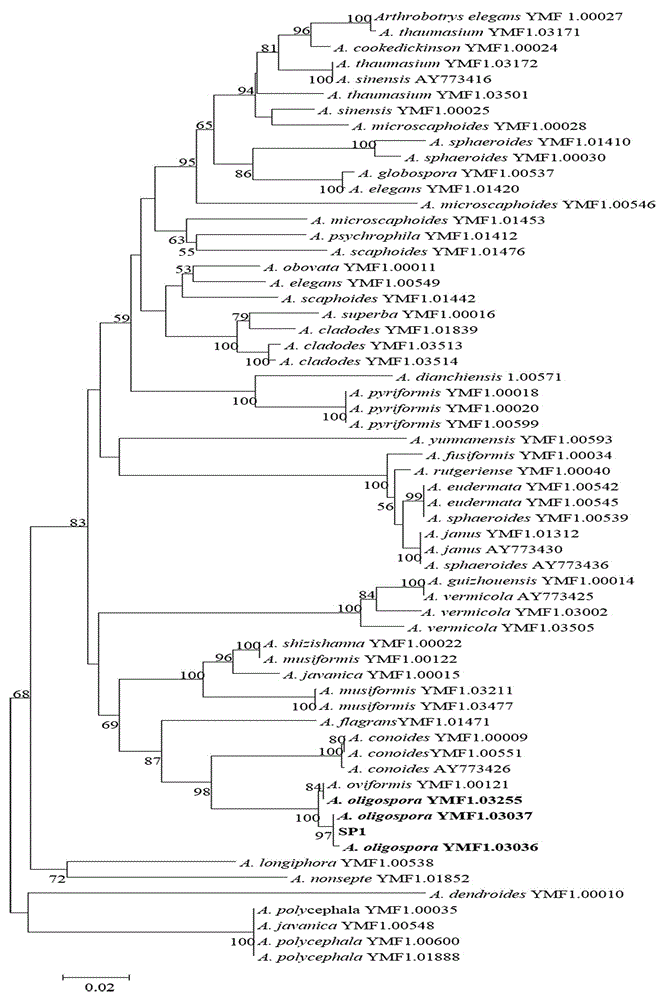
[0029] (1) The suspected but unidentified Arthroplexia pure culture strain SP1 was selected and divided into two parts, one was identified by traditional taxonomic methods, and the other was identified by the method used in the present invention.
[0030] (2) Using the steps described in Example 1 to extract the genomic DNA in the strain, using SEQ ID NO.55 and SEQ ID NO.56 as primers to amplify the RPB2 gene sequence fragment. As shown in Example 1, PCR product detection, sequencing, and DNA barcode sequence acquisition were carried out, and then the following results were judged.
[0031] (3) By comparing the sequence of the fungus to be identified with the existing sequence in the gene bank, the genetic distance method based on Kimura-2-parameter probability and the phylogenetic tree method based on cluster analysis are used to establish identification rules. Identify species and obtain the similari...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com