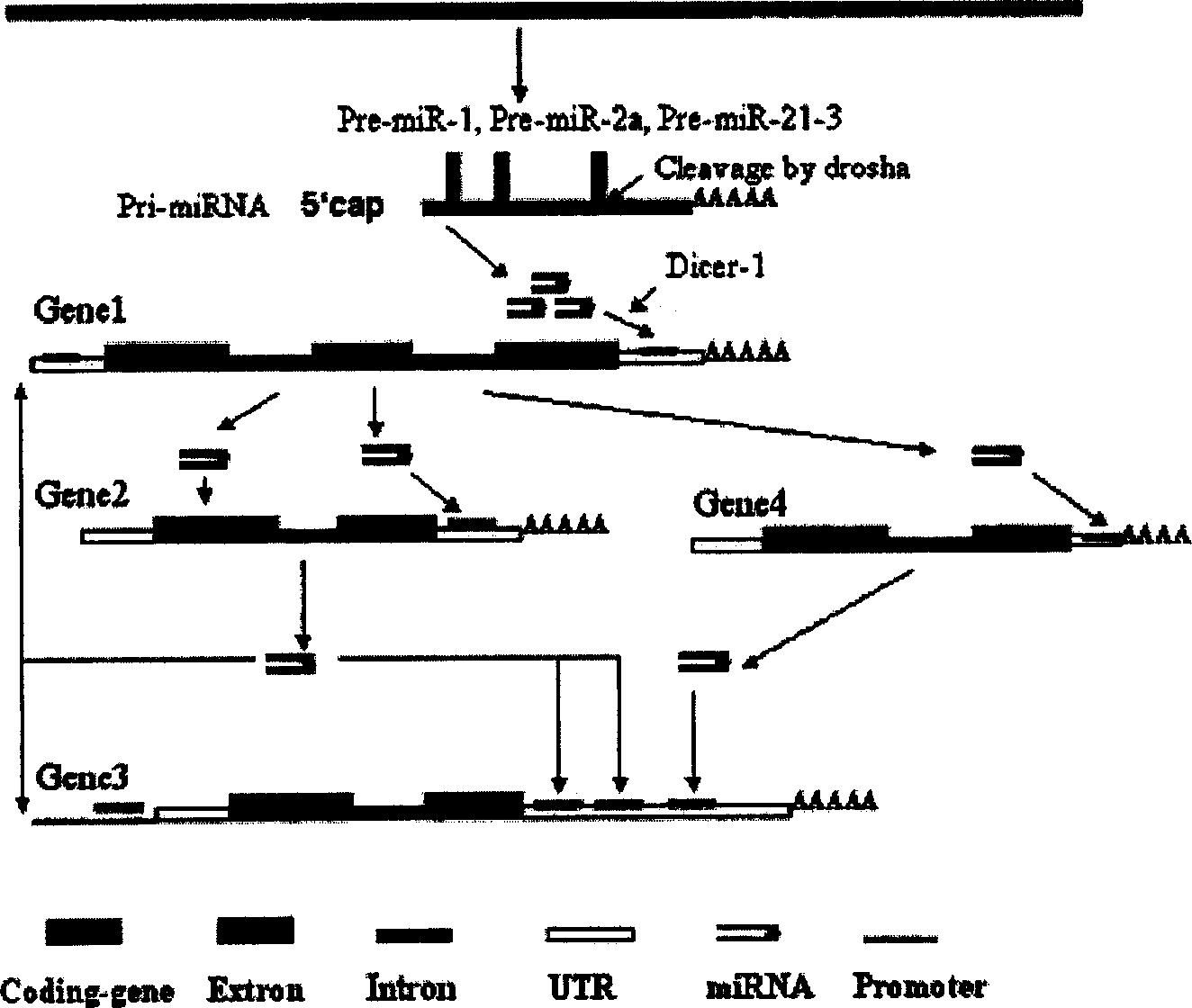
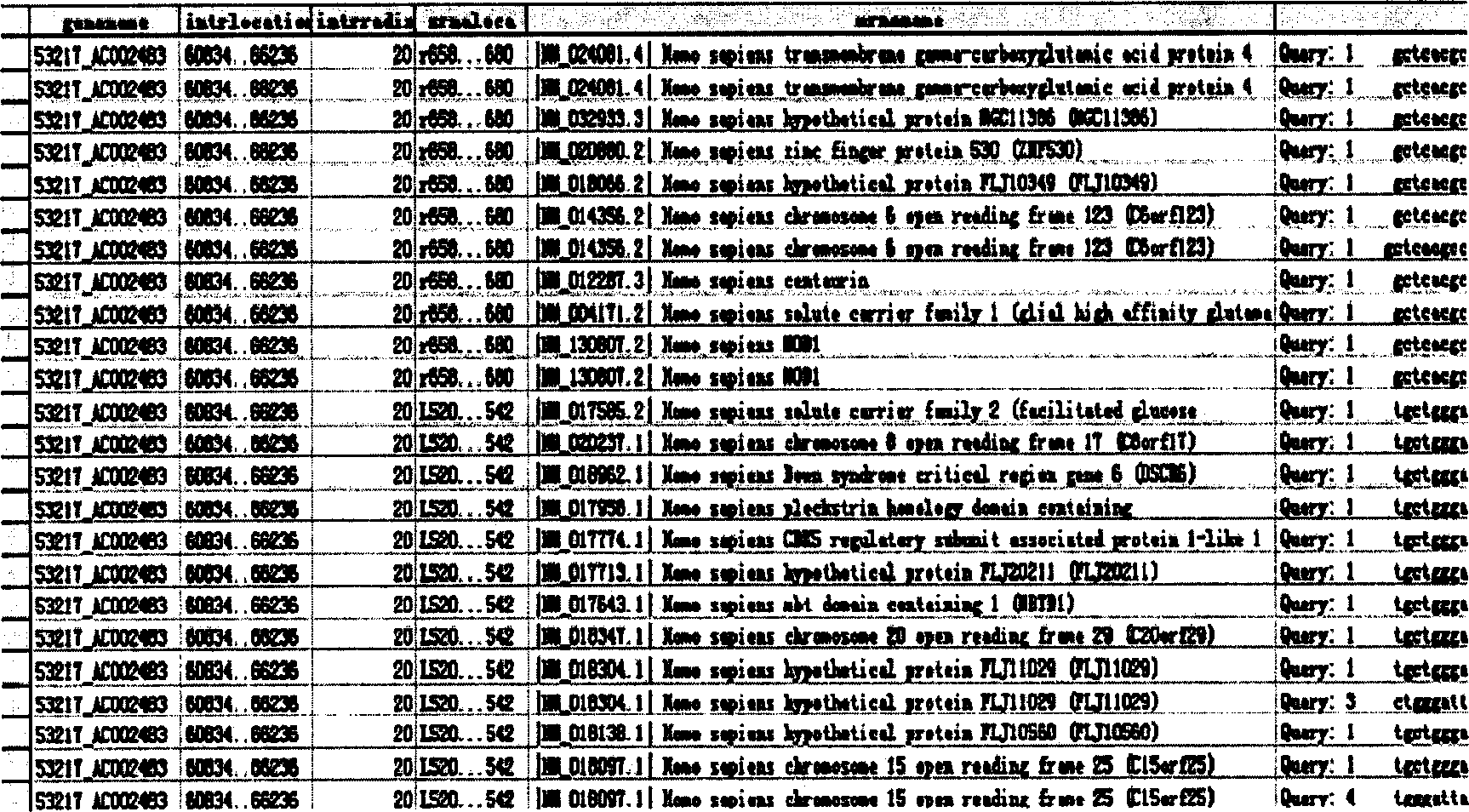
Human autogenous siRNA sequence, its application and screening method
A DNA sequence, endogenous technology, applied in the field of bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0013] 1. Terminology
[0014] In the context of the present invention, the term "double-stranded oligonucleotide" as used is a duplex of polymers or oligomers containing nucleotides. Such as a double-stranded RNA molecule (dsRNA), a double-stranded DNA molecule (dsDNA), a double-stranded sRNA-cDNA hybrid molecule. The term further includes oligonucleotides composed of natural nucleotides, sugars and covalent linkages between nucleotides, as well as modified or non-natural nucleotides. Each of these oligomer types and their numerous derivatives have been extensively reported. Those modified or substituted nucleotides are often superior to natural nucleotides. If they are used to synthesize corresponding oligonucleotides, their products may have stronger enzymatic resistance and better uptake by cells performance and higher affinity performance with its target nucleic acid.
[0015] As used herein, the terms "double-stranded RNA molecule (dsRNA), double-stranded DNA molecule...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com