Taq DNA polymerase mutant and application thereof
A polymerase, mutant technology, applied in the biological field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1 The acquisition of four kinds of mutants
[0032] According to the conventional method, use Nanjing Nuoweizan Biotechnology Co., Ltd. Max Master Mix (P515) and Ultra One Step Cloning Kit (C115) performs site-directed mutation on Taq DNA polymerase (sequence shown in SEQID NO.1) to obtain mutants, which are named: Mut1, Mut2, Mut3 and Mut4.
[0033] The mutation sites of Mut1 are: T386A, A407L, F413Y (sequence shown in SEQ ID NO.2);
[0034] The mutation sites of Mut2 are: T386A, A407L (sequence shown in SEQ ID NO.3);
[0035] The mutation sites of Mut3 are: A407L, F413Y (sequence shown in SEQ ID NO.4);
[0036] The mutation sites of Mut4 are: T386A, F413Y (sequence shown in SEQ ID NO.5).
[0037] The primers used for Mut1 point mutation are shown in Table 1-2 below:
[0038] Table 1 Mut1 primer sequence
[0039] Primer name 5'-3' sequence 1-1F CCAACACCGCCCCCGAGGGGGTG 1-1R TCGGGGGCGGTGTTGGAAGGGTCCAG 2-1F GCCCTCCTTTCCGAGAG...
Embodiment 2 4
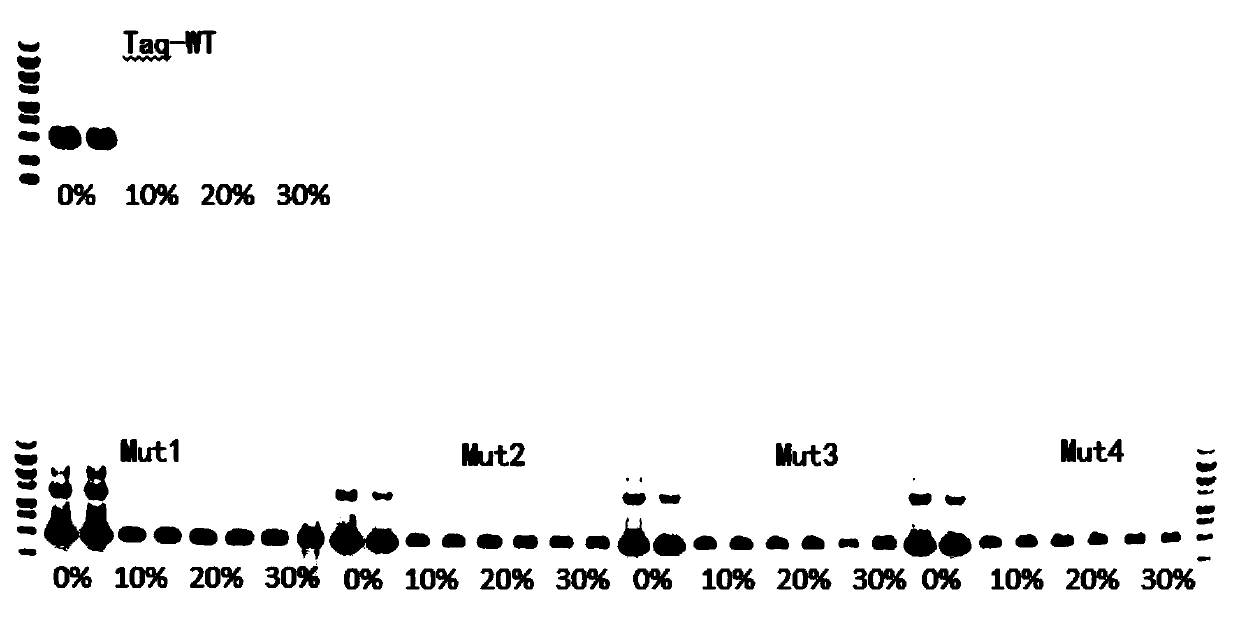
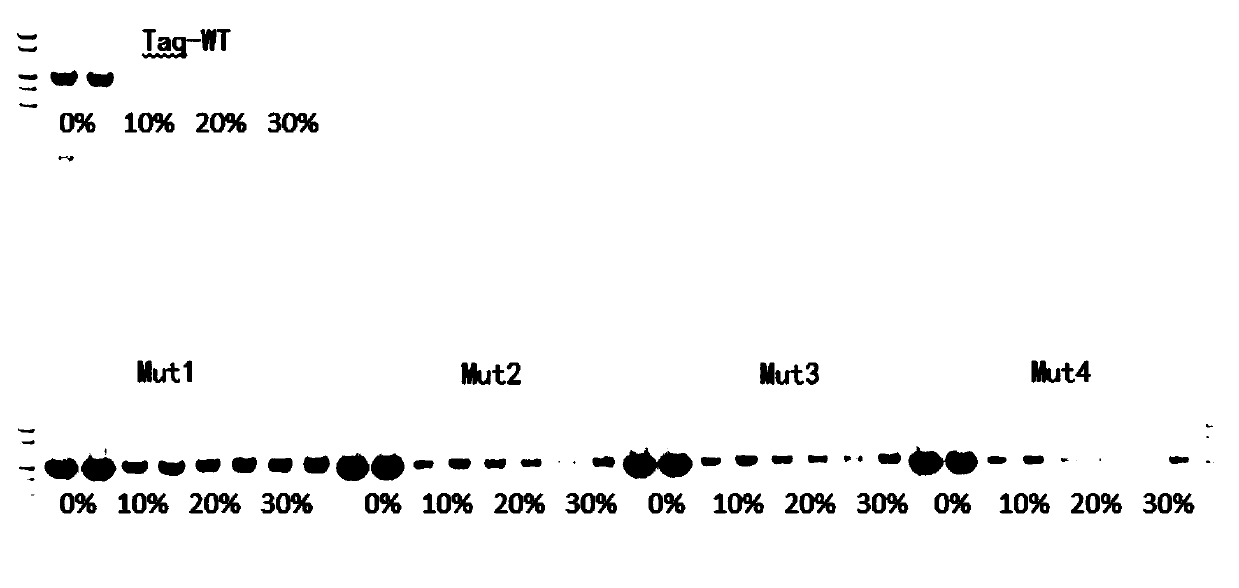
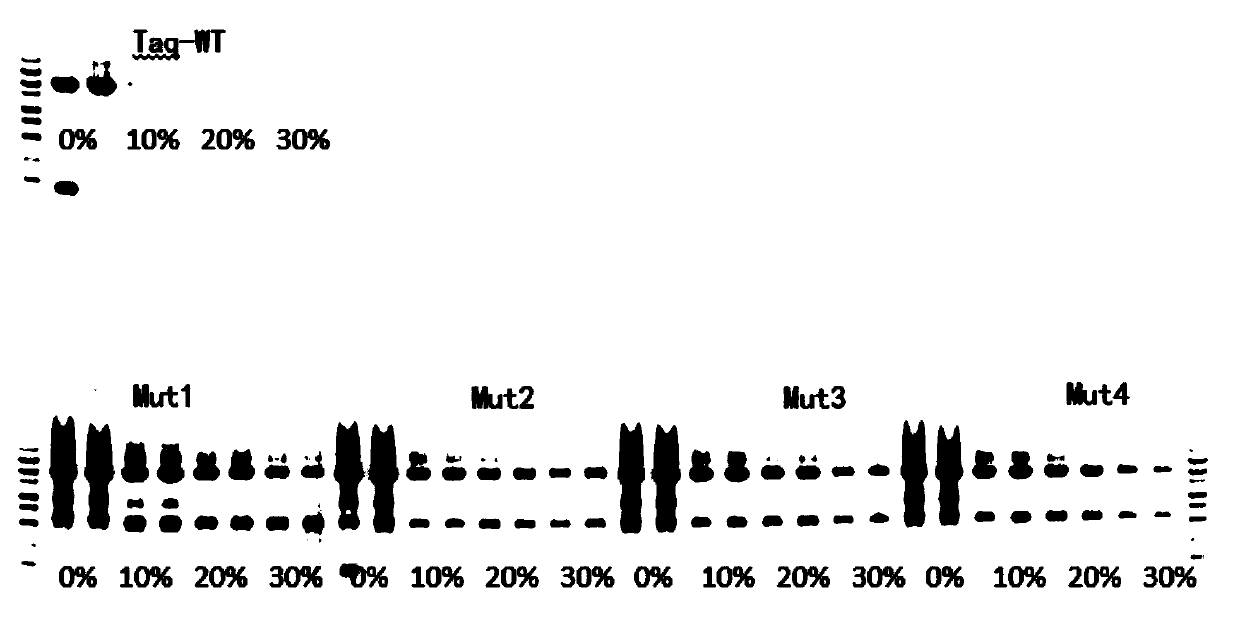
[0062] Example 2 Four kinds of mutant Taq have higher blood tolerance
[0063] The wild-type Taq and the four mutants were prepared into 2×PCR Mix according to the following formula, and five kinds of 2×PCR Mix were obtained.
[0064] 2×PCR Mix1: 200mM Tris-HCl, 100mM KCl, 0.8mM dNTP, 4mM MgCl 2 , 0.2U / μl Taq;
[0065] 2×PCR Mix2: 200mM Tris-HCl, 100mM KCl, 0.8mM dNTP, 4mM MgCl 2 , 0.2U / μl Mut1;
[0066] 2×PCR Mix3: 200mM Tris-HCl, 100mM KCl, 0.8mM dNTP, 4mM MgCl 2 , 0.2U / μl Mut2;
[0067] 2×PCR Mix4: 200mM Tris-HCl, 100mM KCl, 0.8mM dNTP, 4mM MgCl 2 , 0.2U / μl Mut3;
[0068] 2×PCR Mix5: 200mM Tris-HCl, 100mM KCl, 0.8mM dNTP, 4mM MgCl 2 , 0.2 U / μl Mut4.
[0069] Five kinds of 2×PCR Mix were mixed according to the following table 20 (50 μl reaction system). In order to control the input amount of template in each well to be consistent, λDNA was used as template and blood was added as impurity to verify blood tolerance.
[0070] Table 9 Mixing method of PCR reaction syst...
Embodiment 3
[0080] Embodiment 3 detects the enzyme activity of four kinds of mutants and wild-type Taq
[0081] The five kinds of 2×PCR Mixes prepared in Example 2 were tested for enzyme activity by conventional methods in the art, and the results are shown in Table 12.
[0082] Table 12 Enzyme activity detection results of 5 kinds of 2×PCR Mix (enzyme activity, unit: mU / μl)
[0083] Enzyme type Repeat one repeat two repeat three average value 2×PCR Mix1 192 190 188 190 2×PCR Mix2 195 183 190 189 2×PCR Mix3 184 191 183 186 2×PCR Mix4 190 187 192 190 2×PCR Mix5 183 197 190 190
[0084] It can be seen from the data in Table 12 that the enzyme activity of the mutant enzyme is not enhanced, but it shows good amplification performance when amplifying blood samples. Therefore, in combination with the results of Example 2, we speculate that: T386A (threonine mutated into alanine), A407L (alanine mutated into leucine), F413Y (phe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com