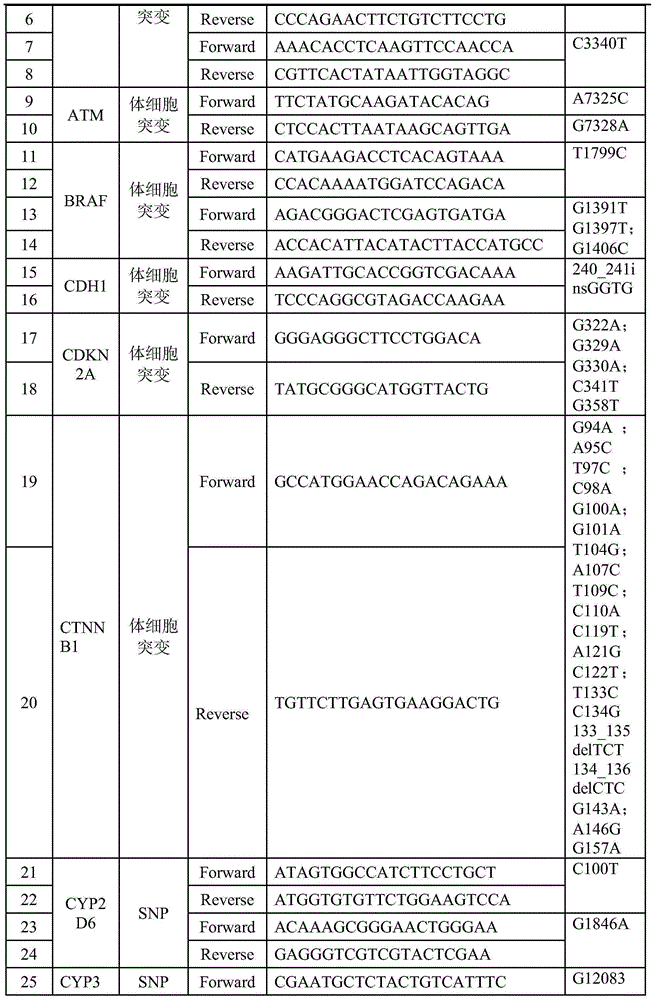
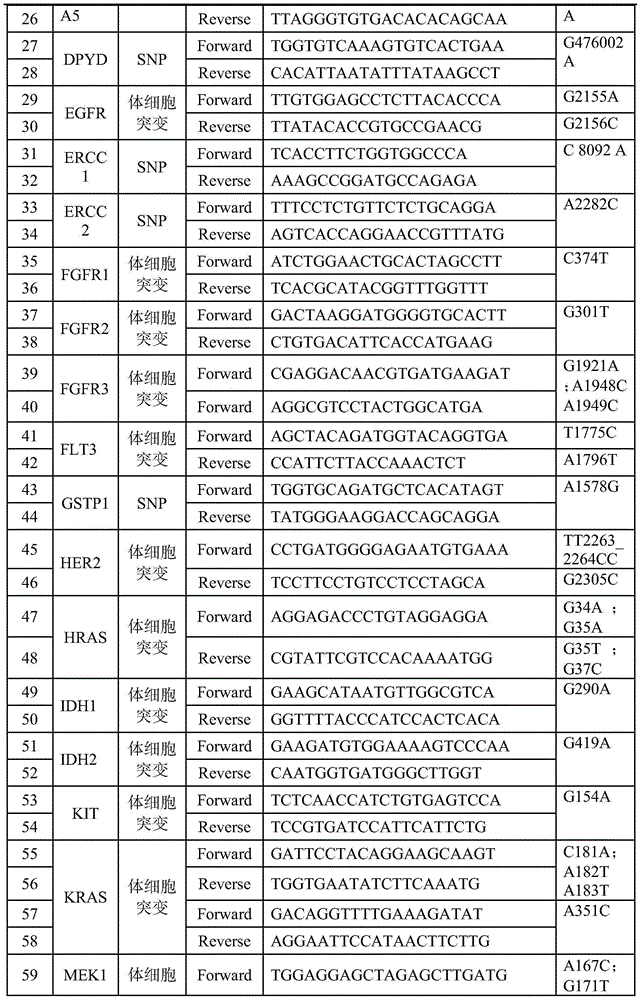
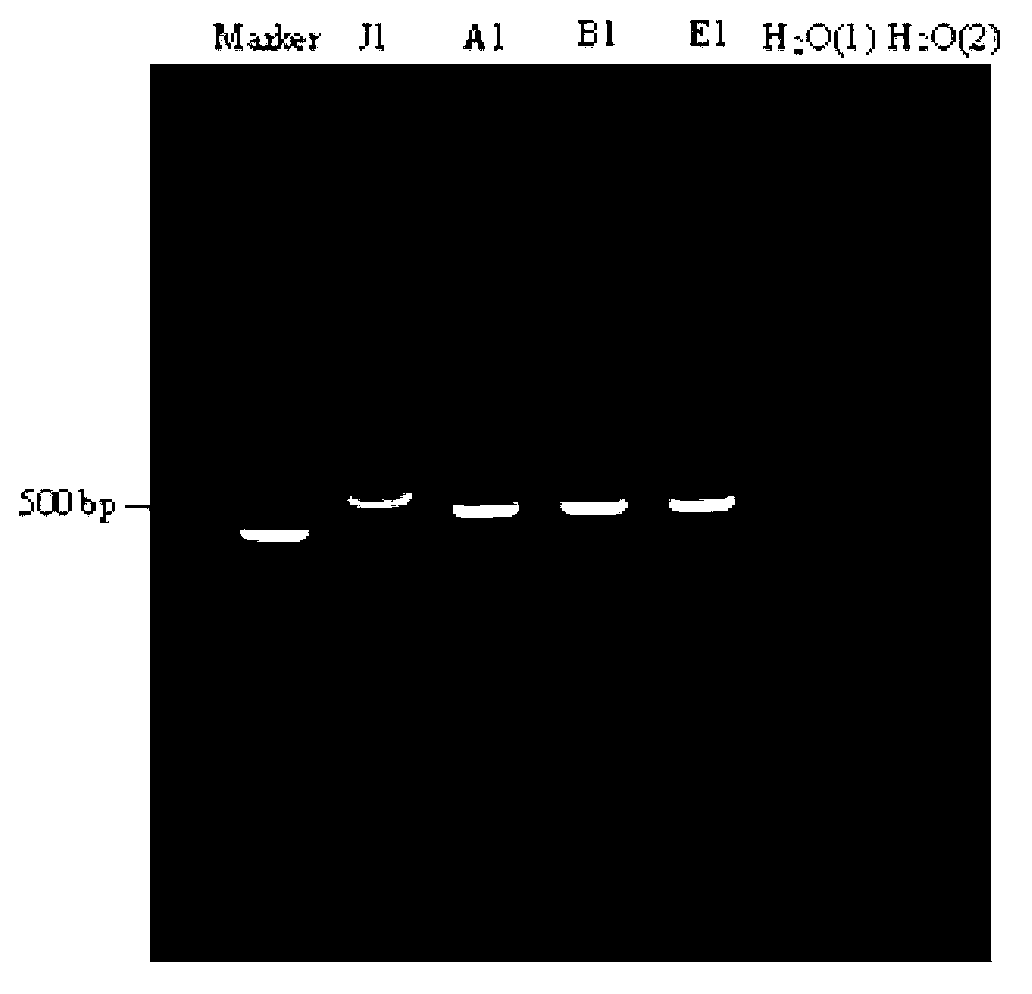Patents
Literature
234results about How to "Good amplification effect" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
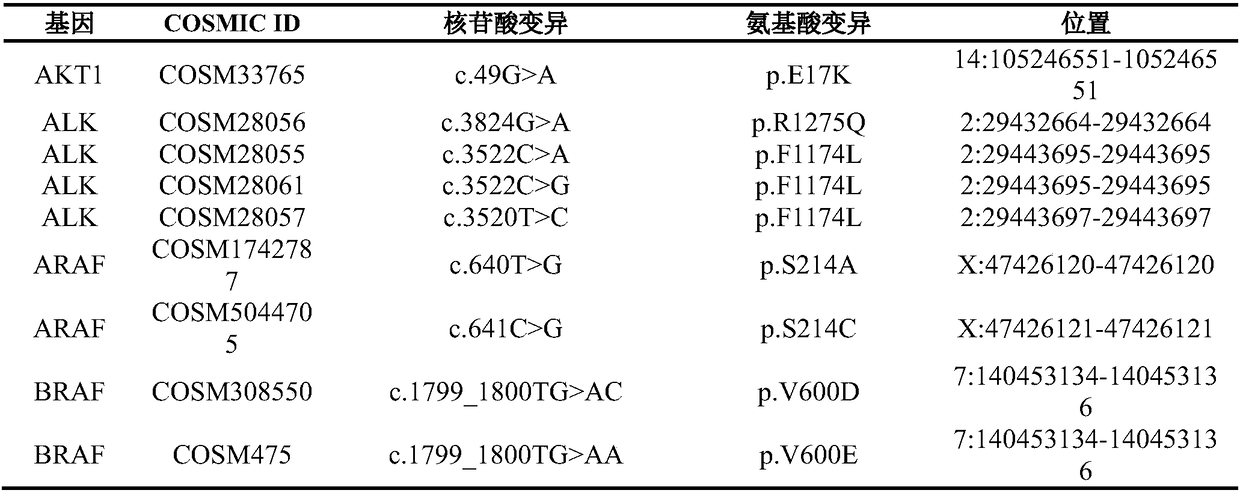
Non-small cell lung cancer targeted therapy gene detection method
InactiveCN105969857AReduce mutual interferenceStrong specificityMicrobiological testing/measurementCell sensitivityOncogene
The invention discloses a non-small cell lung cancer targeted therapy gene detection method, and belongs to the field of gene detection. The method for detecting 466 mutations of 12 oncogenes is developed by multiplex PCR and high throughput sequencing technologies, wherein the oncogenes are AKT1, ALK, BRAF, EGFR, ERBB4, FGFR1, FGFR2, FGFR3 , KRAS, MET, PIK3CA, and PTEN, and the mutations may be substitutions, insertions and / or deletions of one or more bases. The detection method provided by the invention has the advantages of high detection sensitivity of up to 0.01%, and clear and objective detection results, can be directly used for reflecting specific mutation sites of the relevant gene, directly used for guiding clinical non-small cell lung cancer targeted dosage, and used for early diagnosis or auxiliary diagnosis and screening of cancers as well as post-cancer surveillance.
Owner:HEFEI INSTITUTES OF PHYSICAL SCIENCE - CHINESE ACAD OF SCI
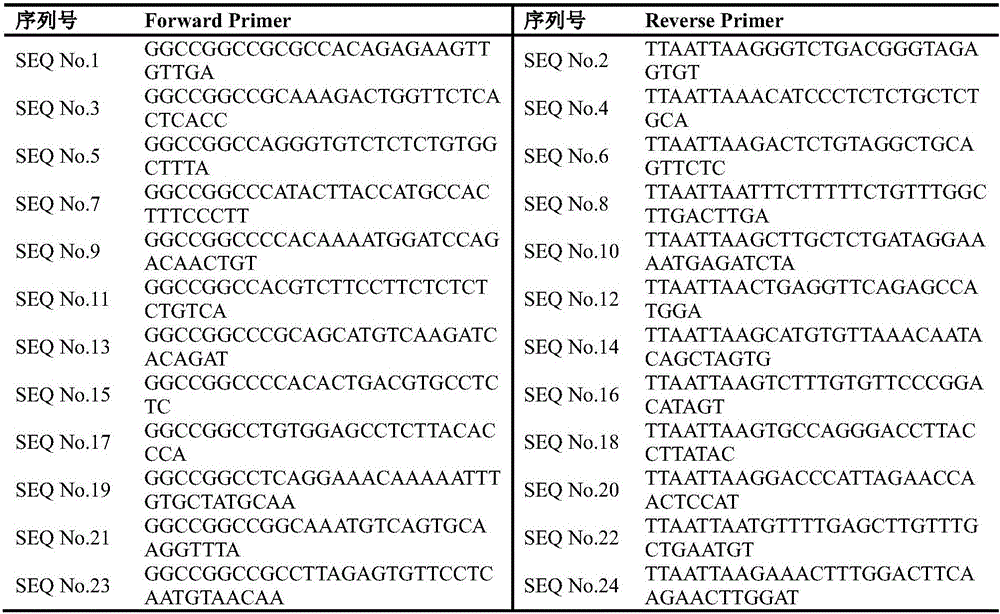
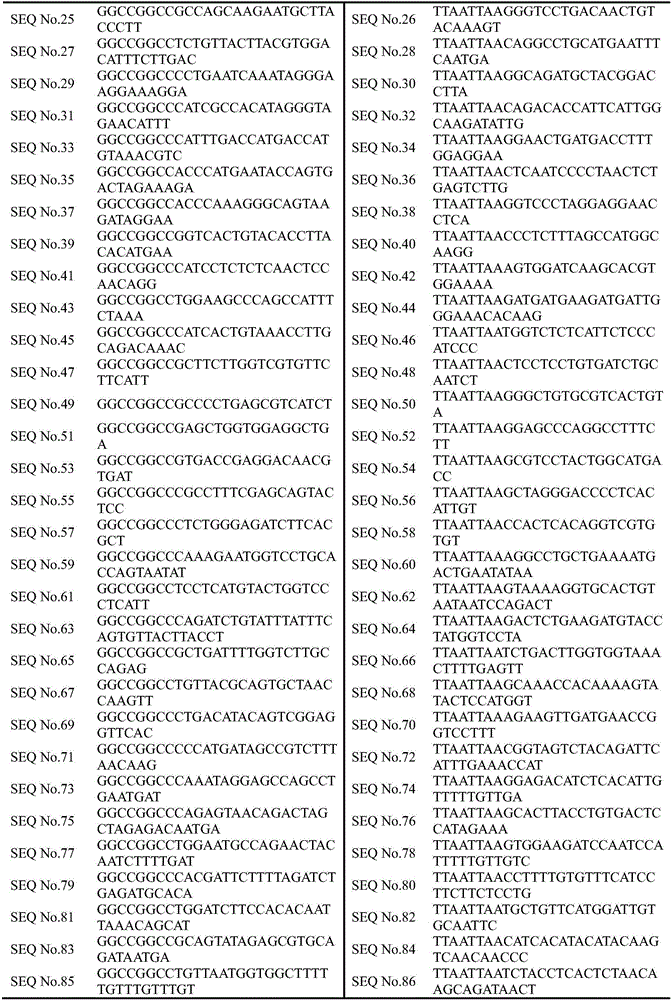
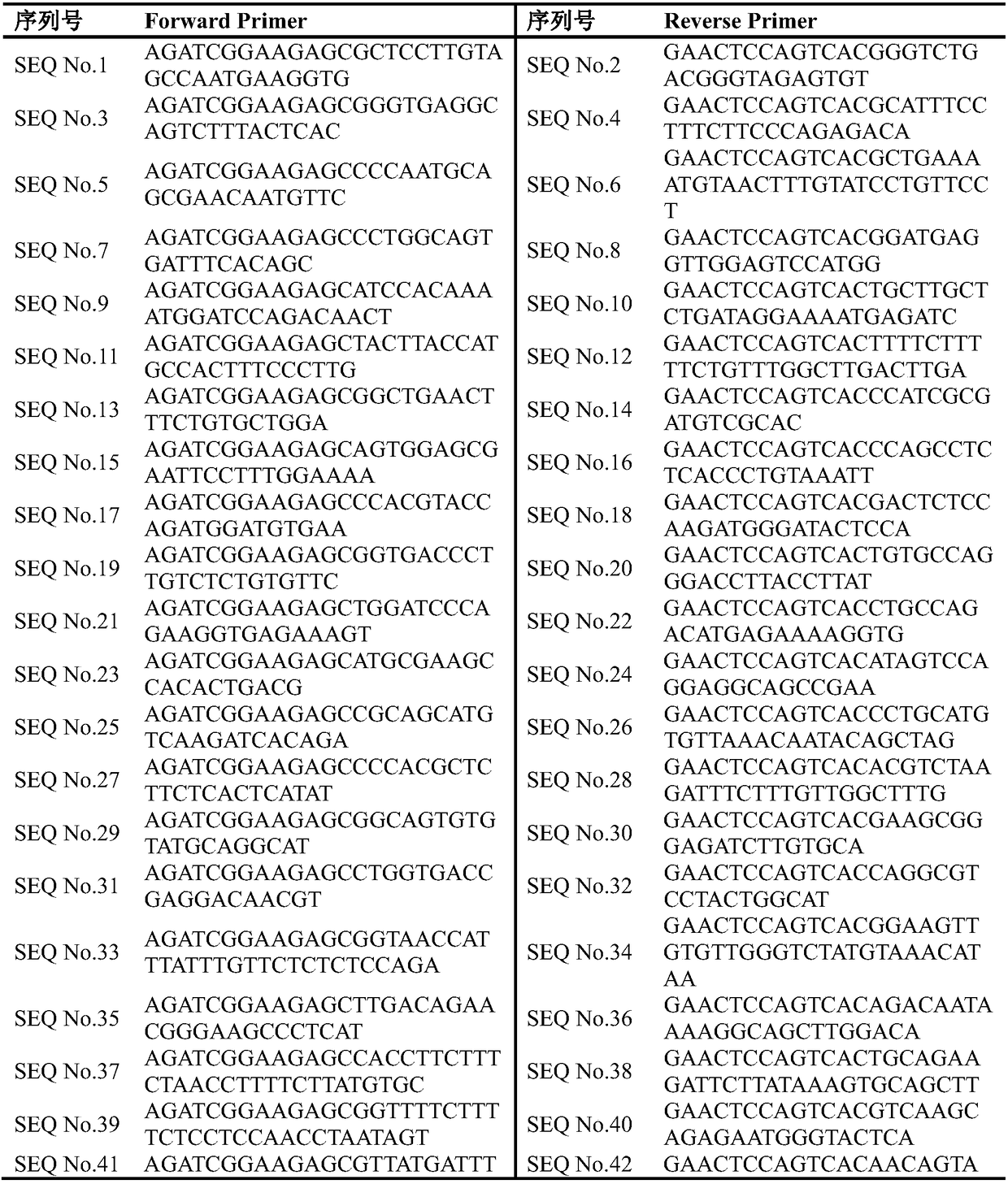
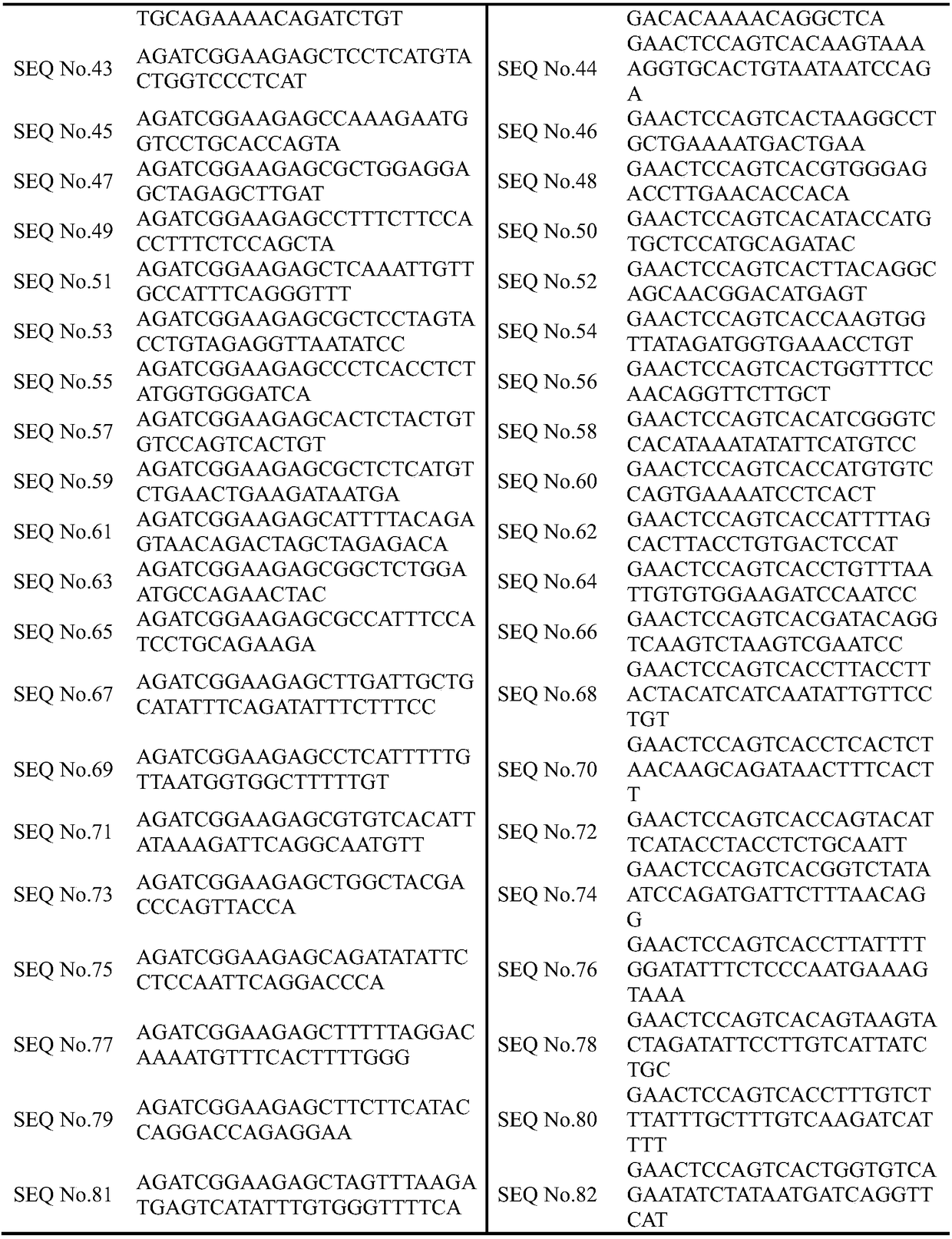
Primer, kit and method for determining lung cancer gene mutation site based on high-flux sequencing technology
InactiveCN108315416AReduce mutual interferenceGood amplification effectMicrobiological testing/measurementDNA/RNA fragmentationWilms' tumorOncogene
The invention discloses a primer, a kit and a method for determining a lung cancer gene mutation site based on a high-flux sequencing technology, which belongs to the field of biological molecular detection. The invention discloses a method and a kit for determining a lung cancer gene mutation site based on a high-flux sequencing technology. The method comprises the following steps: extracting tumor tissue DNA; designing a panel of a lung cancer targeting treatment molecular diagnosis associated gene; performing the PCR primer amplification; and establishing a library, and performing the high-flux sequencing. The specific primer and the kit for determining the lung cancer gene mutation site are used for detecting 316 mutation situations of 16 oncogenes, and the mutation can be the replacement, insertion and / or deletion of one or more alkaline groups. The sensitivity of the detection method and the kit of the invention can reach up to 1 percent, the detection result is definite and objective and can directly reflect the specific mutation site of the reaction associated gene and has important significance for early diagnosis or auxiliary diagnosis and screening of the cancer and theprognosis monitoring of the cancer.
Owner:HEFEI INSTITUTES OF PHYSICAL SCIENCE - CHINESE ACAD OF SCI +1
Pre-amplification method for trace DNA applied in medicolegal expertise
InactiveCN102634510AMeet the needs of identificationHigh DNA samplesMicrobiological testing/measurementDNA preparationDNABioinformatics
The invention discloses a pre-amplification method for trace DNA applied in medicolegal expertise. The method aims at amplifying the trace DNA to obtain high-concentricity DNA template so as to facilitate following medicolegal expertise. The method mainly comprises the steps of exploring the optimum concentration and proportion volume of combined primers; and the amplification conditions are further optimized, the trace DNA aims for being amplified with high efficiency and high fidelity, so that the possibility for the next legal medical expert is provided for determining individuals.
Owner:KUNMING UNIV OF SCI & TECH
Kit for simultaneously detecting nucleic acids of seven respiratory pathogens and application thereof
ActiveCN110964854AEasy to degradeLower requirementMicrobiological testing/measurementMicroorganism based processesRNA extractionMycoplasma pneumoniae pneumonia
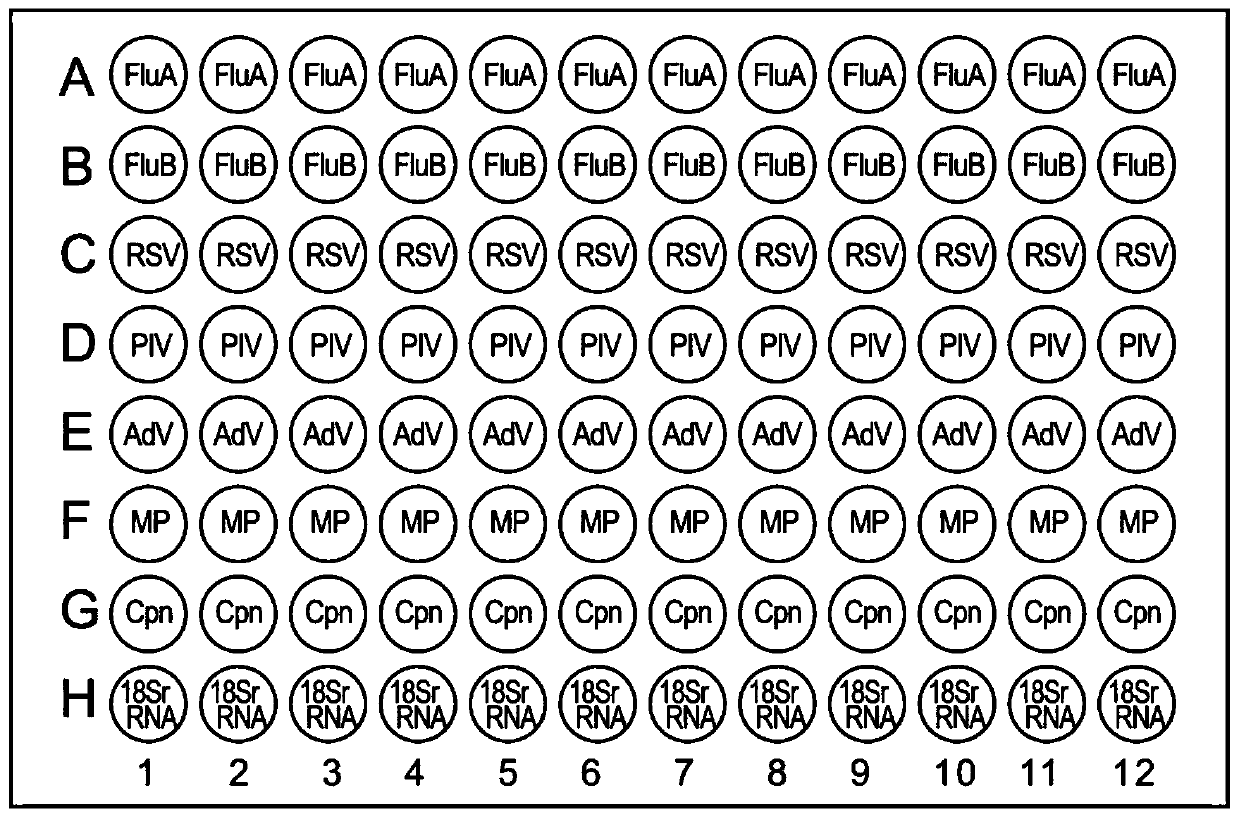
The invention discloses a kit for simultaneously detecting nucleic acids of seven respiratory pathogens and application of the kit. The kit is based on a double amplification technology (RNA isothermal amplification and multi-biotin signal amplification), and can detect H1N1 / H3N2 type influenza A viruses, influenza B viruses, respiratory syncytial viruses, 1 / 2 / 3 type human parainfluenza viruses, B / E type adenoviruses, mycoplasma pneumoniae and chlamydia pneumoniae. The kit of the invention does not need RNA extraction, and is not prone to pollution, high in sensitivity and good in specificityin the process of detection, and can be widely applied to detection of the nucleic acids of the above seven respiratory pathogens.
Owner:武汉中帜生物科技股份有限公司
Specific primer pair based on rbcL gene and used for identifying land plant species and application thereof
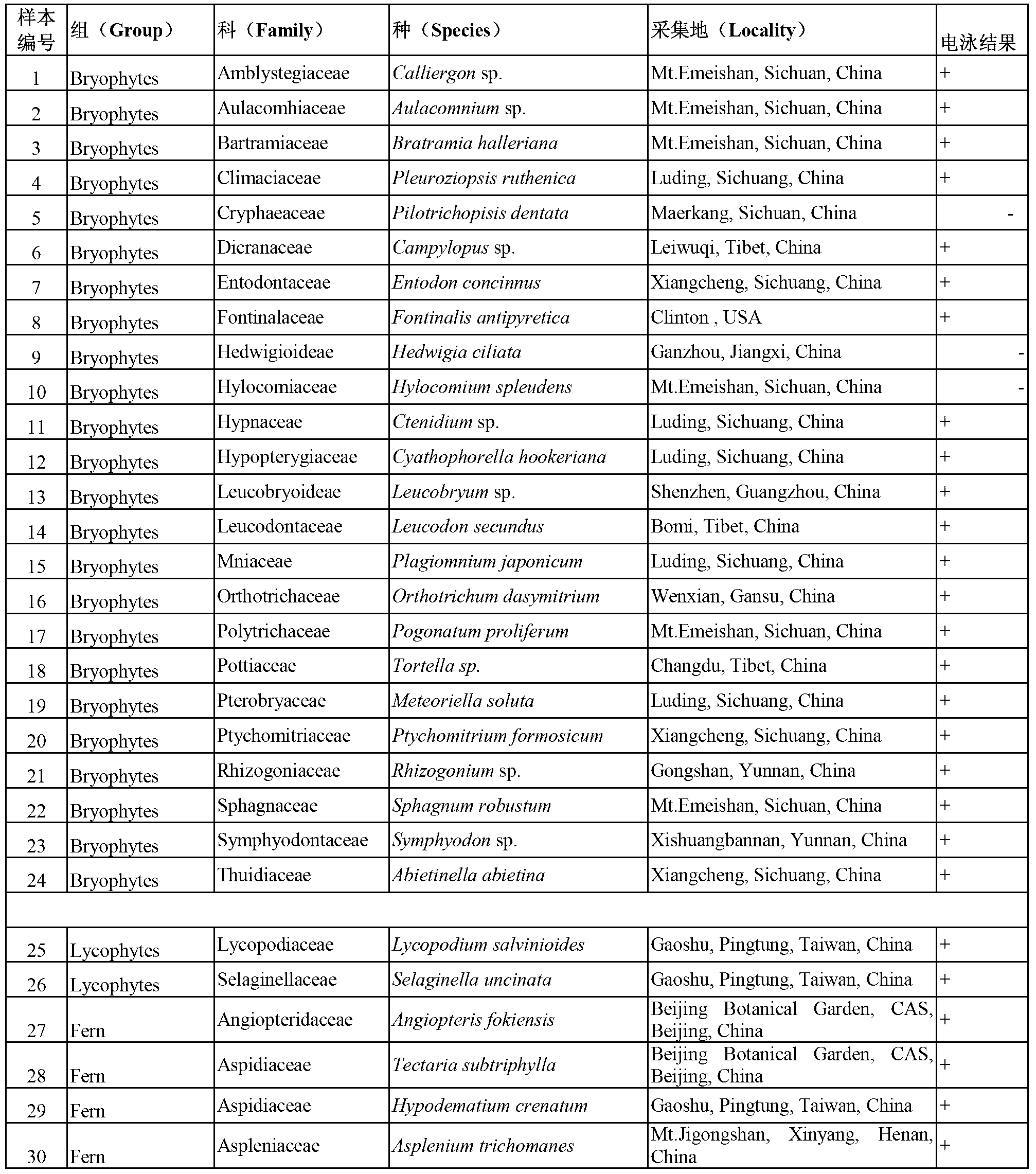
InactiveCN103305620AGood amplification effectEfficient identificationMicrobiological testing/measurementDNA/RNA fragmentationDNA barcodingDNA fragmentation
The invention discloses a specific primer pair based on a rbcL gene and used for identifying land plant species and an application thereof. The specific primer pair provided by the invention is composed of a single stranded DNA A and a single stranded DNA B, wherein the single stranded DNA A is 15-40bp and comprises a DNA fragment of the DNA fragments as shown by a sequence 1 of a sequence table; and the single stranded DNA B is 15-40bp and comprises a DNA fragment of the DNA fragments as shown by a sequence 2 of the sequence table. The single stranded DNA A can be the DNA fragment as shown by the sequence 1 of the sequence table; and the single stranded DNA B can the DNA fragment as shown by the sequence 2 of the sequence table. The specific primer pair provided by the invention can be used for developing a universal kit, effectively identifying the land plant species and prompting the development and social service of plant DNA bar code.
Owner:INST OF BOTANY CHINESE ACAD OF SCI
Mixed type hot-start DNA polymerase composition, PCR amplification kit, and applications thereof
ActiveCN108546749AImprove heat resistanceLow mismatch rateMicrobiological testing/measurementBiotechnologyWild type
The invention discloses a mixed type hot-start DNA polymerase composition, a PCR amplification kit, and applications thereof. The composition comprises Taq DNA polymerase, an anti-Taq antibody, and mutant type KOD DNA polymerase. The anti-Taq antibody is capable of specifically being combined with Taq DNA polymerase. The 147th site of wild type KOD DNA polymerase is histidine, and the 147th site of mutant type KOD DNA polymerase is lysine. The experiment results show that the mismatch rate is low when the mixed type hot-start DNA polymerase composition is used for PCR amplification, the composition has the dual characteristics of high efficient amplification and high fidelity and is heat resistant in amplification, and the amplification effect is good no matter for a long DNA segment, a template with a low amount, or a template with a high GC content.
Owner:深圳市艾伟迪生物科技有限公司
Nucleic acid, kit and method for detecting G1165C polymorphism of human ADRB1 gene
InactiveCN106520979AEasy to operateOvercome pollutionMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceNon specific
The invention discloses nucleic acid and a kit for detecting G1165C polymorphism of the human ADRB1 gene, and also establishes a method for detecting the G1165C polymorphism of the human ADRB1 gene, which is strong in specificity, high in sensitivity, high in degree of accuracy, and easy to operate. The kit for detecting the G1165C polymorphism of the human ADRB1 gene is applicable to the selection for various clinical samples, and has the remarkable advantages of strong specificity, high sensitivity, short experimental period, operation simplicity, safety and non-toxicity, low cost, and the like. The detection method provided by the invention adopts the completely closed tube operation, and is easy, convenient and rapid to operate, the detection result is obtained by directly directing fluorescence signal values during the PCR process, the PCR aftertreatment or electrophoresis detection is not needed, the large possibility of pollution and false positive property during the conventional PCR technology are overcome, the difficulty of non-specific amplification can be effectively avoided, and the detection method is suitable for detection samples in large batches.
Owner:武汉海吉力生物科技有限公司
Culture system and method for amplifying hematopoietic stem cells and/or hematopoietic progenitor cells, hematopoietic stem cells, and hematopoietic progenitor cells
ActiveCN108235708AGood amplification effectSafe for clinical useBlood/immune system cellsCell culture active agentsMolecular biologyHematopoietic stem cell migration
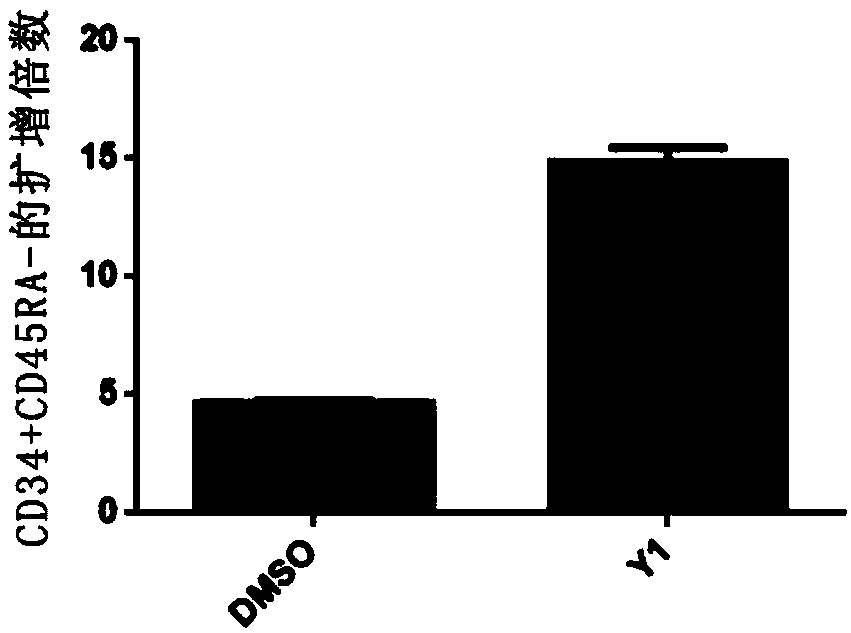
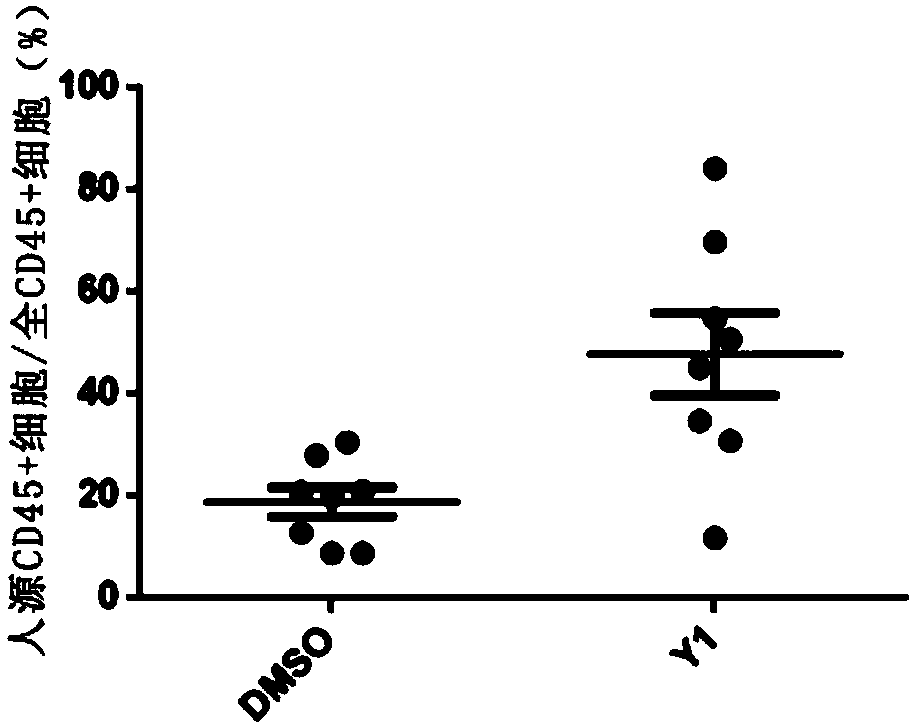
The present invention relates to a culture system and method for amplifying hematopoietic stem cells and / or hematopoietic progenitor cells, hematopoietic stem cells, and hematopoietic progenitor cells. The culture system comprises: a basal medium suitable for stem cell expansion; and a JNK signaling pathway inhibitor. The culture system provided by the invention can expand hematopoietic stem cellsin vitro, and the amplification effect thereof is obvious, and the CD34+CD45RA-labeled hematopoietic stem cells can be amplified nearly 100 times.
Owner:NEWISH TECH (BEIJING) CO LTD
Fusion type Taq DNA polymerase as well as preparation method and application thereof
ActiveCN111690626AHigh affinityImprove bindingMicrobiological testing/measurementTransferasesReverse transcriptaseWild type
The invention provides fusion type Taq DNA polymerase as well as a preparation method and application thereof. The fusion type Taq DNA polymerase comprises a Taq DNA polymerase mutant and DNA bindingprotein, wherein the Taq DNA polymerase mutant is protein obtained by carrying out substitution mutation on one or more sites of a helix-hairpin-helix DNA binding region of wild type Taq DNA polymerase. The fusion type Taq DNA polymerase has enhanced nucleic acid binding capacity, amplification speed and impurity tolerance, has good resistance to inhibition of reverse transcriptase, and has good amplification performance for trace templates, and a biological sample can be directly detected without a nucleic acid extraction step; and a constructed one-step RT-qPCR kit is high in detection speed, high in sensitivity and good in accuracy, and has a wide application prospect in the field of RNA virus detection.
Owner:VAZYME BIOTECH NANJING
Fluorescent constant-temperature amplification technique
The invention discloses a fluorescent constant-temperature amplification technique. In water used as solvent for a fluorescent constant-temperature amplification reagent used in the technique, Tris-buffer solution, magnesium acetate, dithiothreitol, betaine, Tween 20, DMSO, mycose, PEG, BSA, dNTPs, and proper amounts of fluorescent dye, polymerase, single strand DNA-binding protein and recombinase are added. A constant-temperature amplification system of the invention is dependent of high-energy molecules such as ATP and creatine phosphate; compared with existing constant-temperature amplification systems, the constant-temperature amplification system is simpler in composition, lower in cost and more convenient to use. The constant-temperature amplification system is capable of amplifying DNA and RNA sequences, is applicable to amplification of complex templates stably and quickly, capable of finishing an amplification reaction within 30min, has high amplification sensitivity and accuracy without rare occurrence of mispairing, and has good amplification effect. The constant-temperature amplification system is applicable to real-time quantification and has a wider range of application.
Owner:GUANGZHOU HEAS BIOTECH CO LTD
Colloidal gold chromatography kit for nucleic acid detection of novel coronaviruses (2019-nCoV) and application thereof
ActiveCN111455099AEasy to degradeAvoid pollutionMicrobiological testing/measurementMicroorganism based processesRNA extractionNucleic acid detection
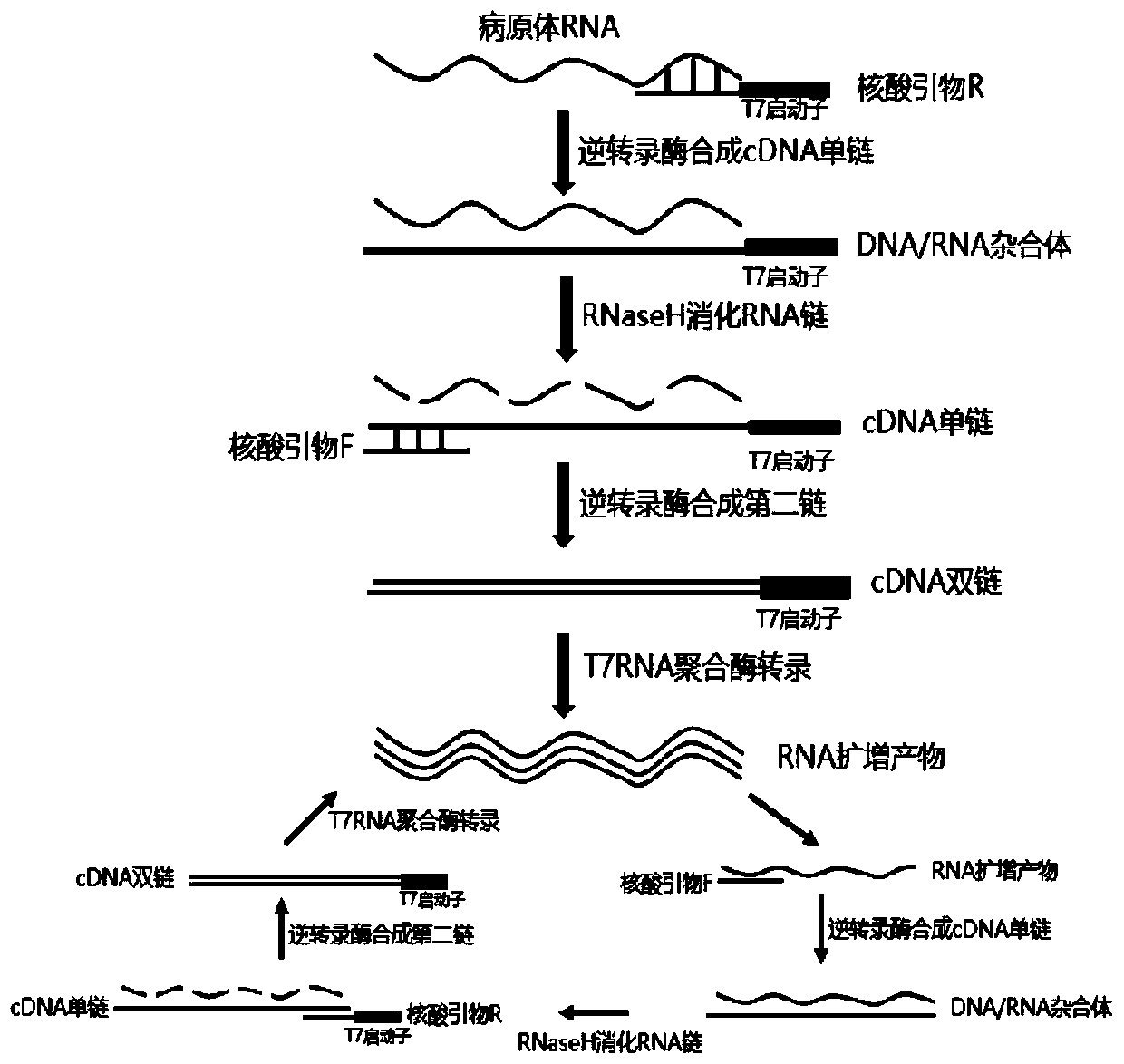
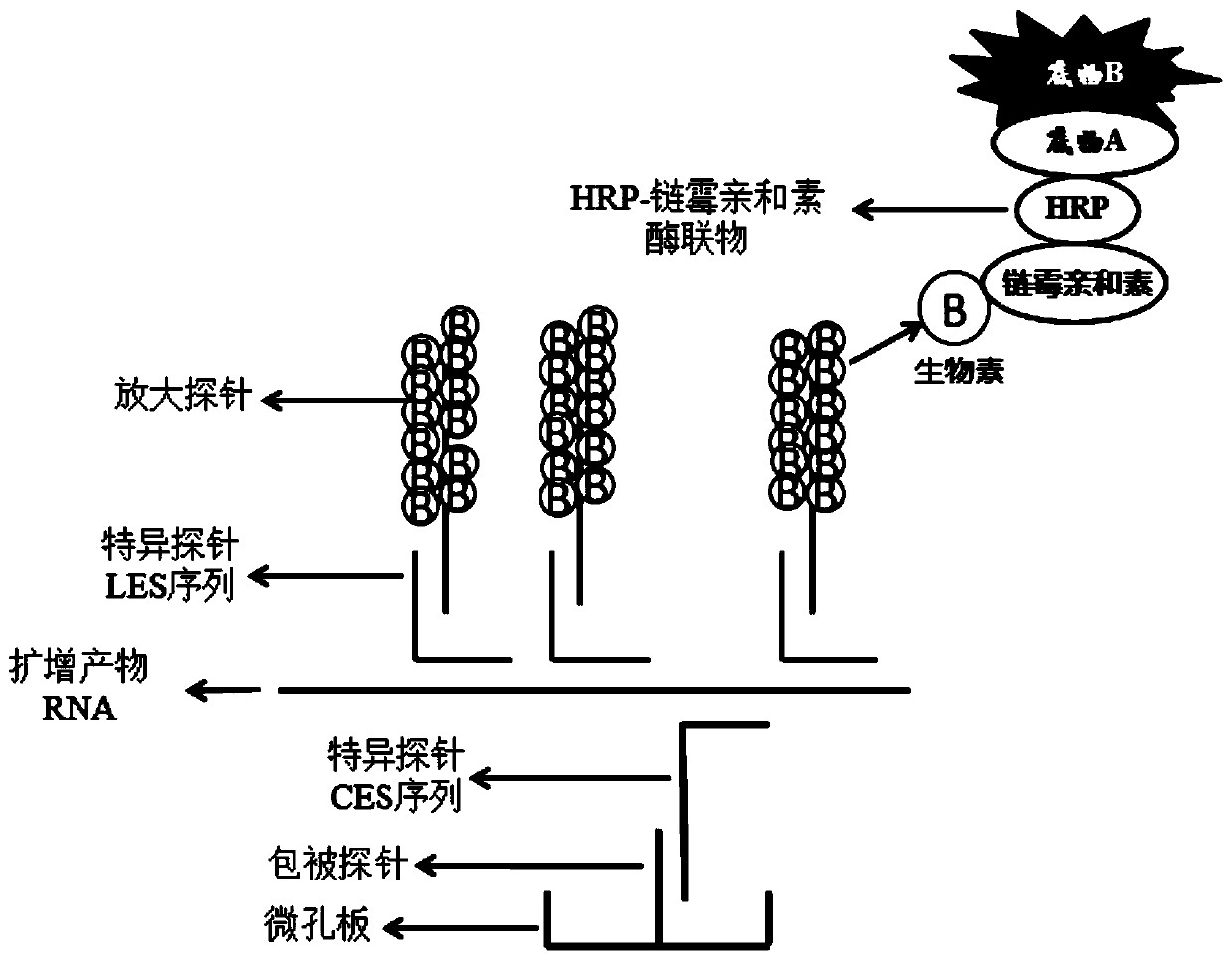
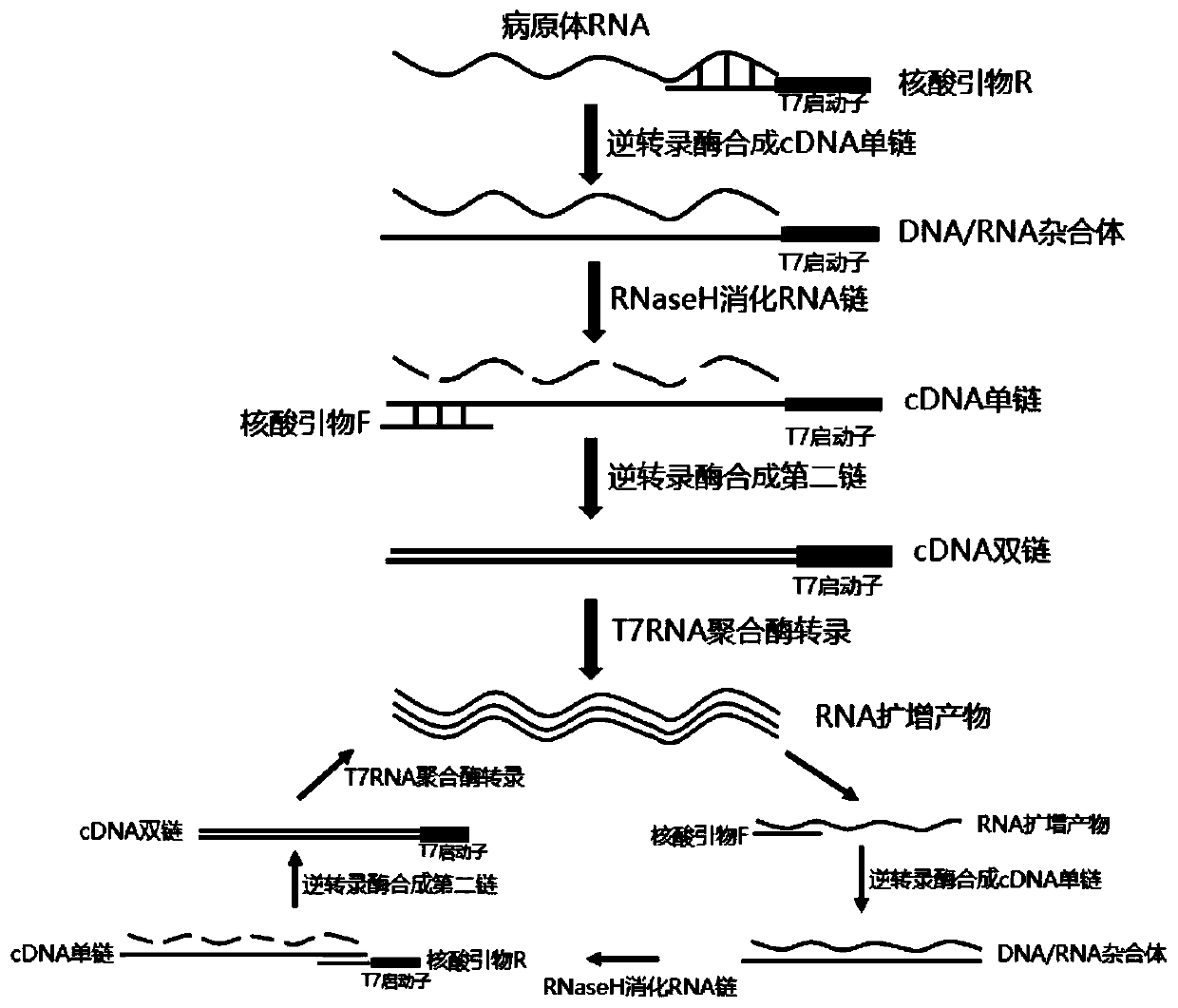
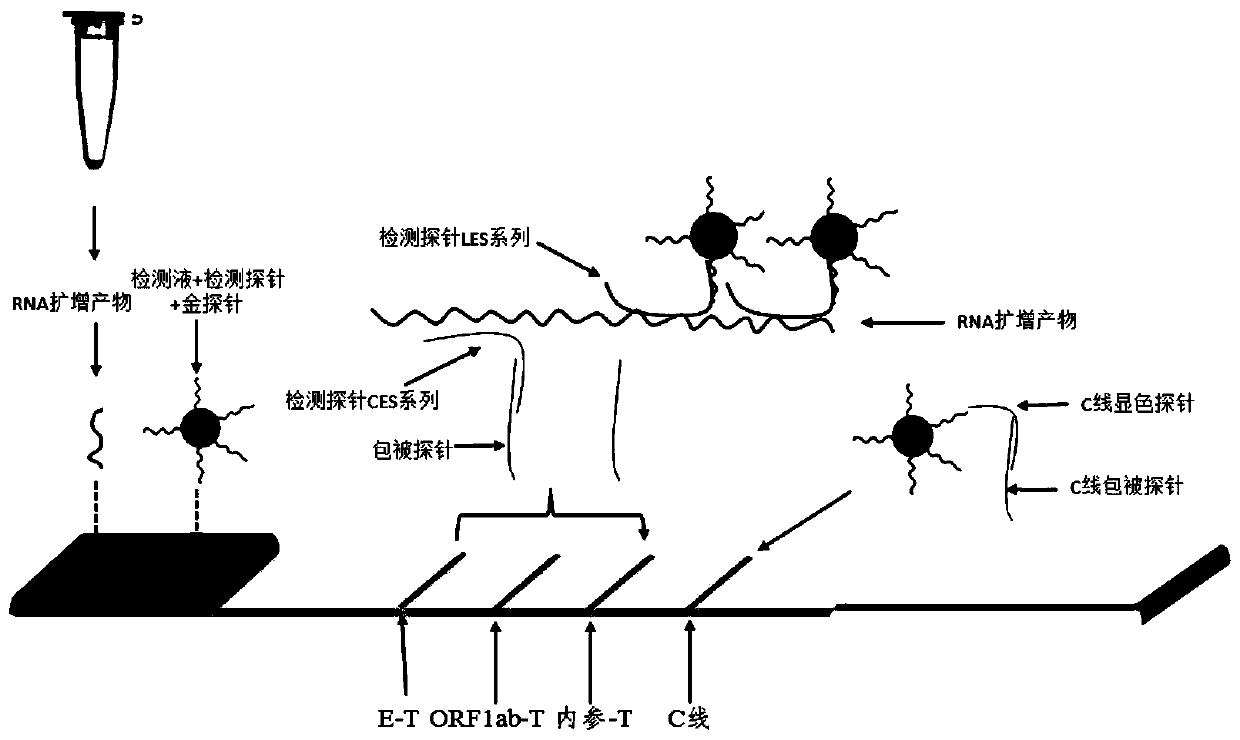
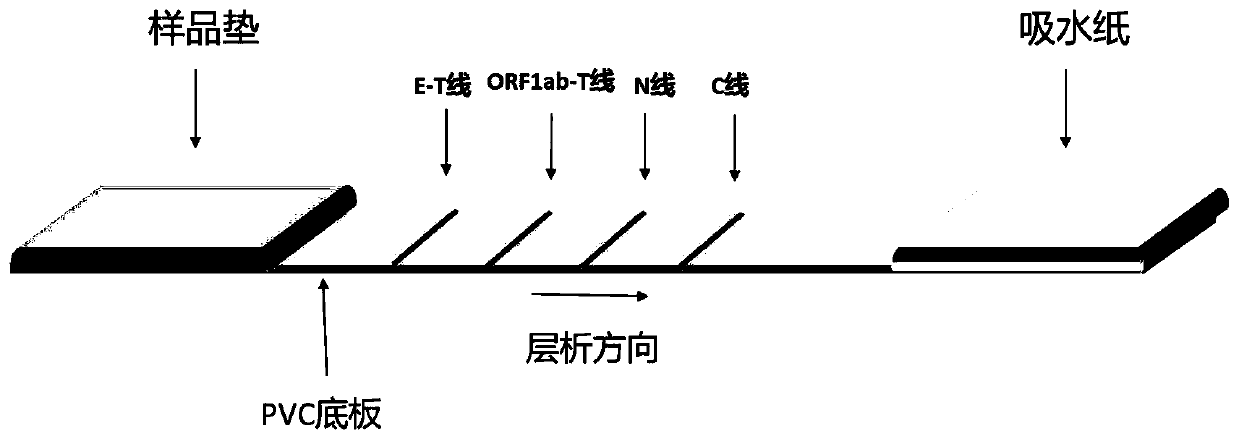
The invention discloses a colloidal gold chromatography kit for detecting novel coronaviruses (2019-nCoV) and an application thereof. According to the kit, a collected sample is cracked by a cell lysis solution to release pathogen nucleic acid, and then under the action of reverse transcriptase and T7RNA polymerase, amplification of pathogen nucleic acid fragments is realized through reverse transcription and transcription processes. An amplified RNA product is recognized and captured by a specific probe in a detection solution to form an RNA amplification product-specific probe-gold probe compound, and the compound is fixed on an NC membrane through lateral flow chromatography to form a visible strip, so that the detection of the pathogen nucleic acid is realized. The kit has no RNA extraction process, does not need a special instrument, has difficulty in pollution in actual detection based on RNA isothermal amplification, has the advantages of high sensitivity, high specificity and simplicity in operation, and makes wide application of novel coronavirus (2019-nCoV) nucleic acid detection possible.
Owner:武汉中帜生物科技股份有限公司
Method for detecting microcystos toxigenicity
InactiveCN1428434AGood reproducibilityHigh sensitivityMicrobiological testing/measurementMicrobiologyMicrocystis
The present invention discloses a method for detecting toxinogeny of microcyst alga. According to the principle of that the microcyst alga toxic synthetase gene mcy B only is existed in toxigenic microcyst alga said invention uses mcyB sequence to design out specific primer TOX1P / 1M and TOX2P / 2M, and utilizes complete cell PCR, preparation of sample, synthesis of primer, PCR reaction and electrophoretic detection to define that the microcyst alga is toxigenic or not. Said invention is short in detection time, good in reproducibility and high in sensibility and accuracy, can be used for massively and quickly identifying microcyst algae in the culture or natural water sample which are toxigenic or not.
Owner:INST OF AQUATIC LIFE ACAD SINICA
Taq DNA polymerase mutant and application thereof
ActiveCN110747183AImprove tolerancePromote amplificationMicrobiological testing/measurementTransferasesAmino acid substitutionBlood specimen
The invention discloses a Taq DNA polymerase mutant and application thereof. The Taq DNA polymerase mutant has amino acid substitutions at one or more of the following amino acid positions in the sequence shown as SEQ ID NO. 1; and the various amino acid substitutions are represented in triplets as 'letters-numbers-letters', wherein the numbers reflect position of an amino acid mutation, the letters before the numbers are corresponding to an amino acid involved in the mutation, and the letters after the numbers reflect an amino acids used to replace the amino acid corresponding to the lettersbefore the numbers. The Taq DNA polymerase mutant disclosed by the invention is capable of changing configuration of an enzyme, thereby improving tolerance of the enzyme; and thus, the mutant can be very well applied in blood sample amplification.
Owner:VAZYME BIOTECH NANJING
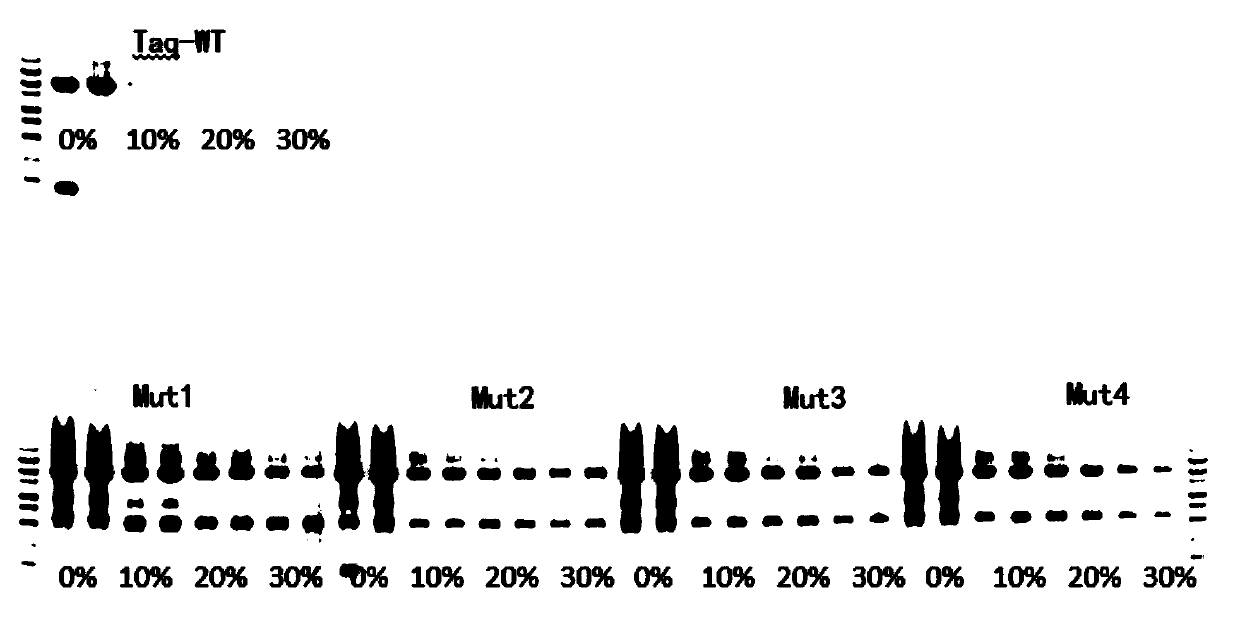
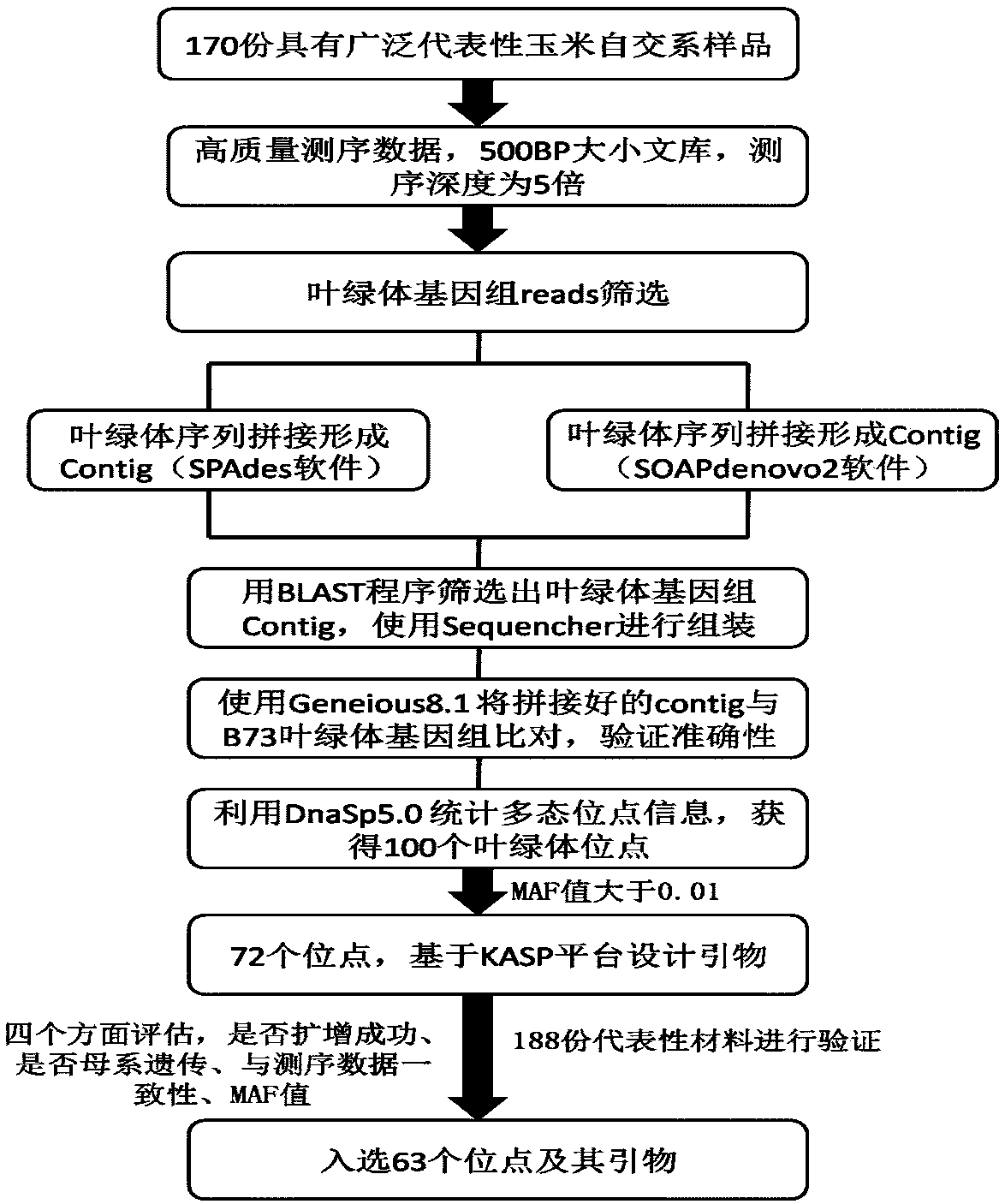
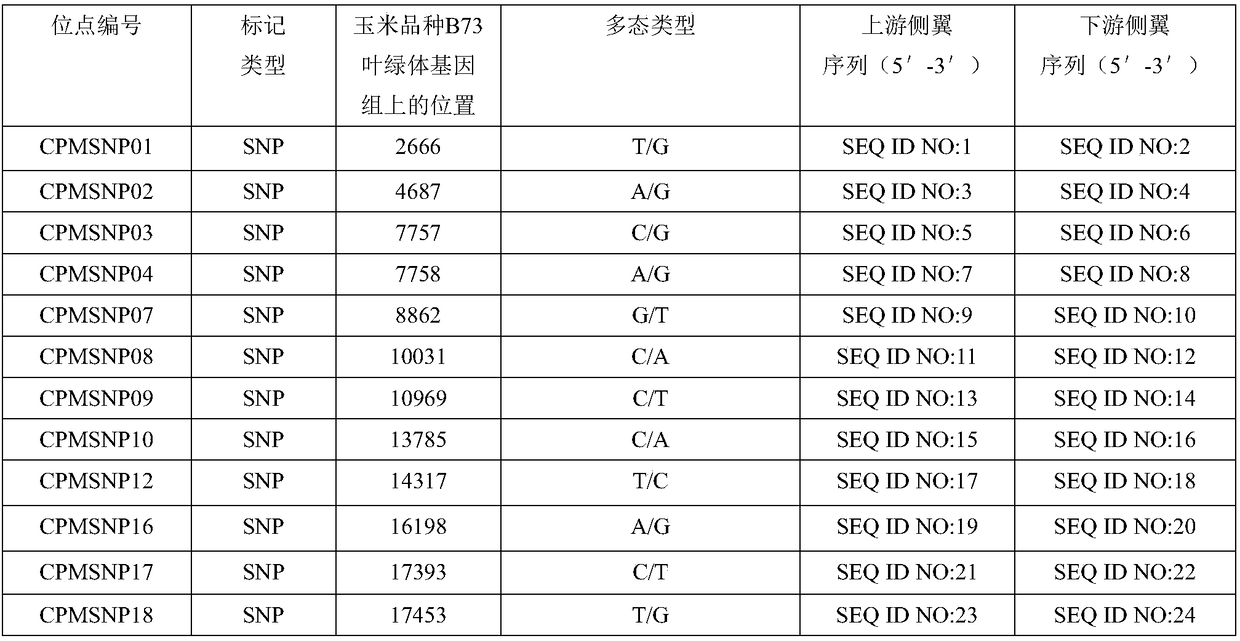
Molecular markers of maize chloroplast genome and its application in cultivar identification
ActiveCN108486266AGuaranteed accuracyImprove efficiencyMicrobiological testing/measurementDNA/RNA fragmentationReciprocal crossGermplasm
The invention provides molecular markerd of a maize chloroplast genome and its application in cultivar identification. The molecular markerd comprise 57 SNP markers and 6 InDel markers. The 63 molecular markers and primers developed according to the markers can be used in any one of the following: (1) constructing a corn variety chloroplast DNA-SNP and InDel fingerprint database, (2) carrying outcorn maternal line traceability analysis, and (3) performing reciprocal cross identification on corn samples. Through the application of the chloroplast polymorphic primer composition, the range of available marker sites of the corn is expanded at the genome level and the new ideas and technical means are provided for the study of maize variety and germplasm resource identification, genetic relationship evaluation and cytoplasmic genetic characteristics.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
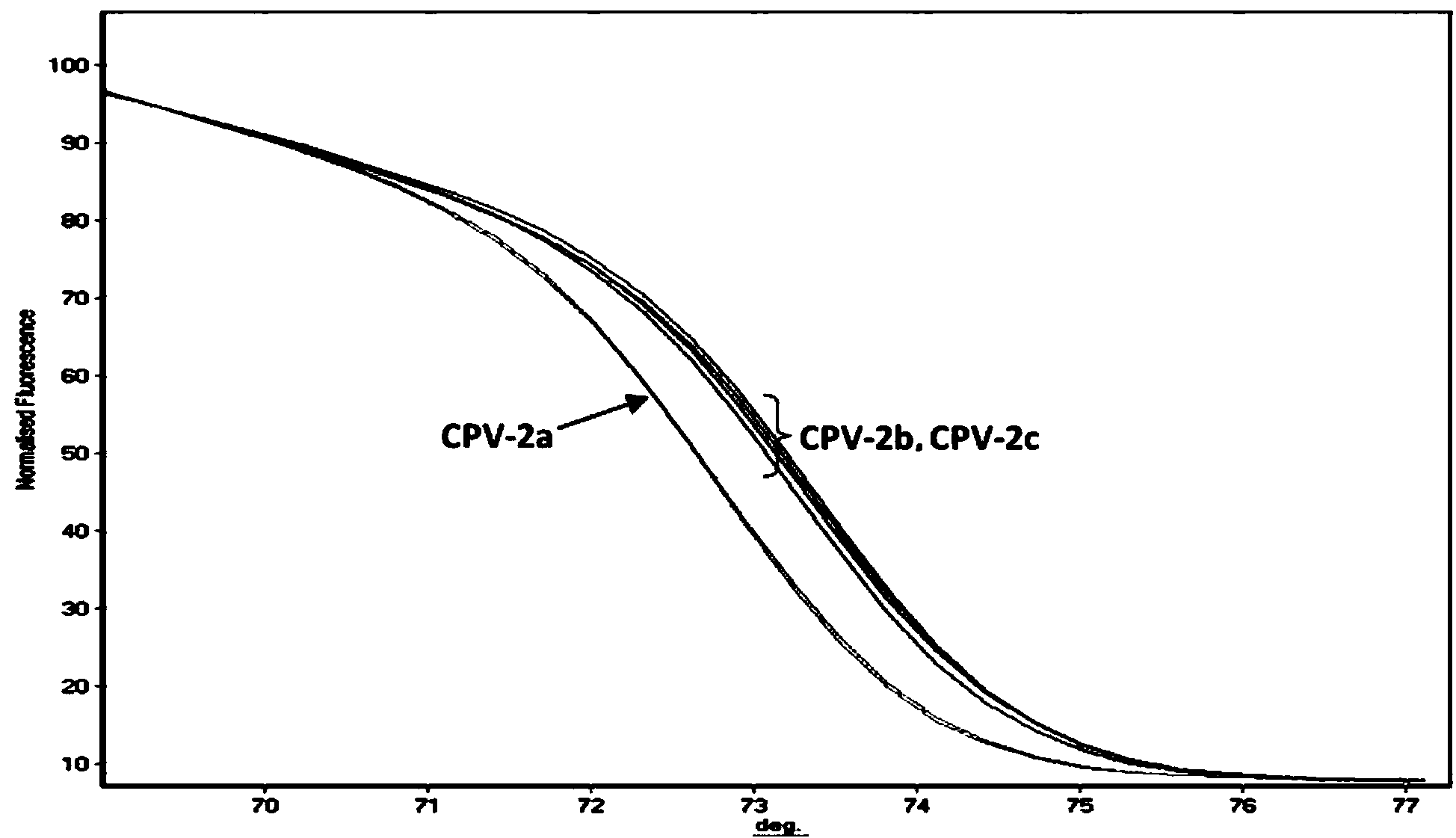
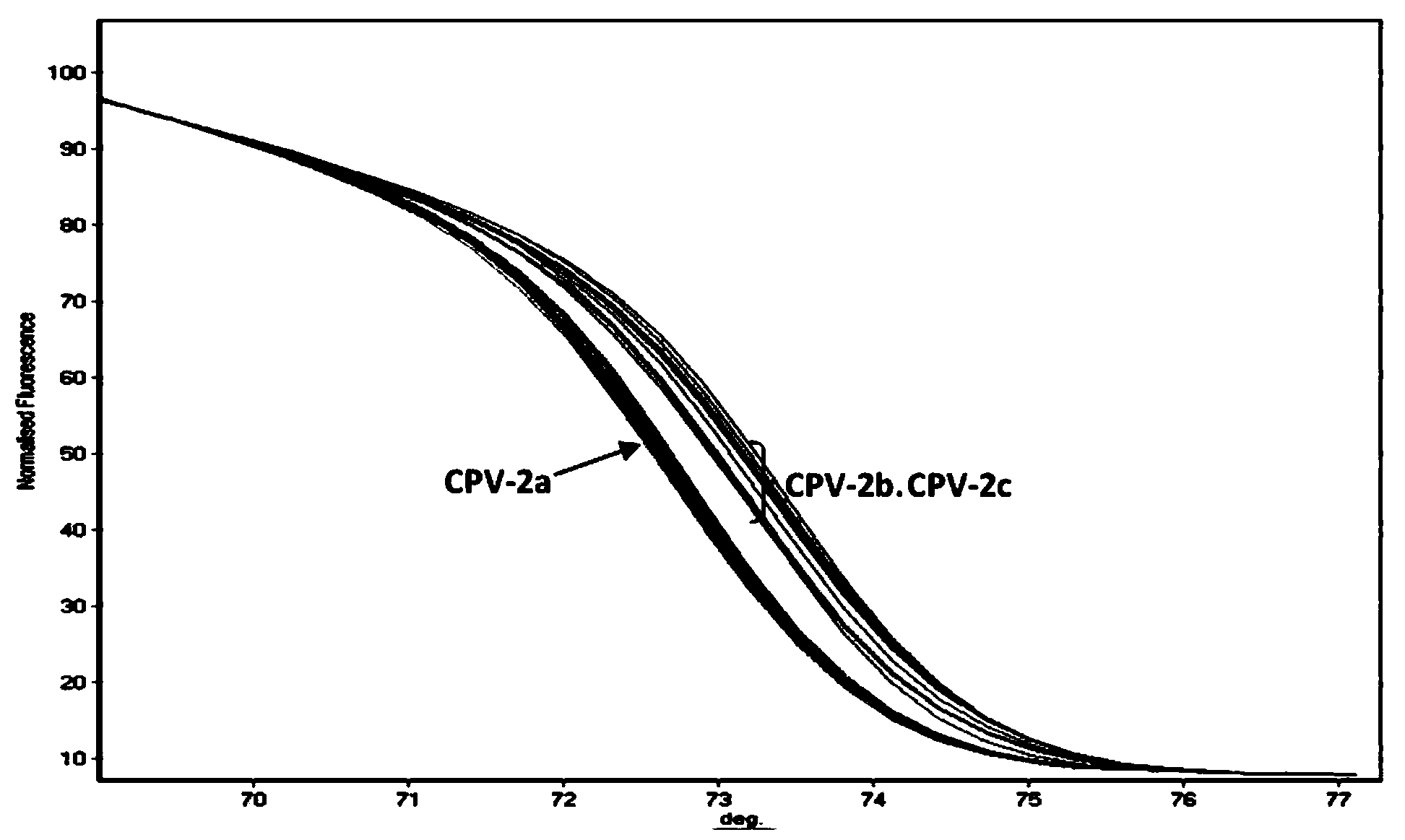
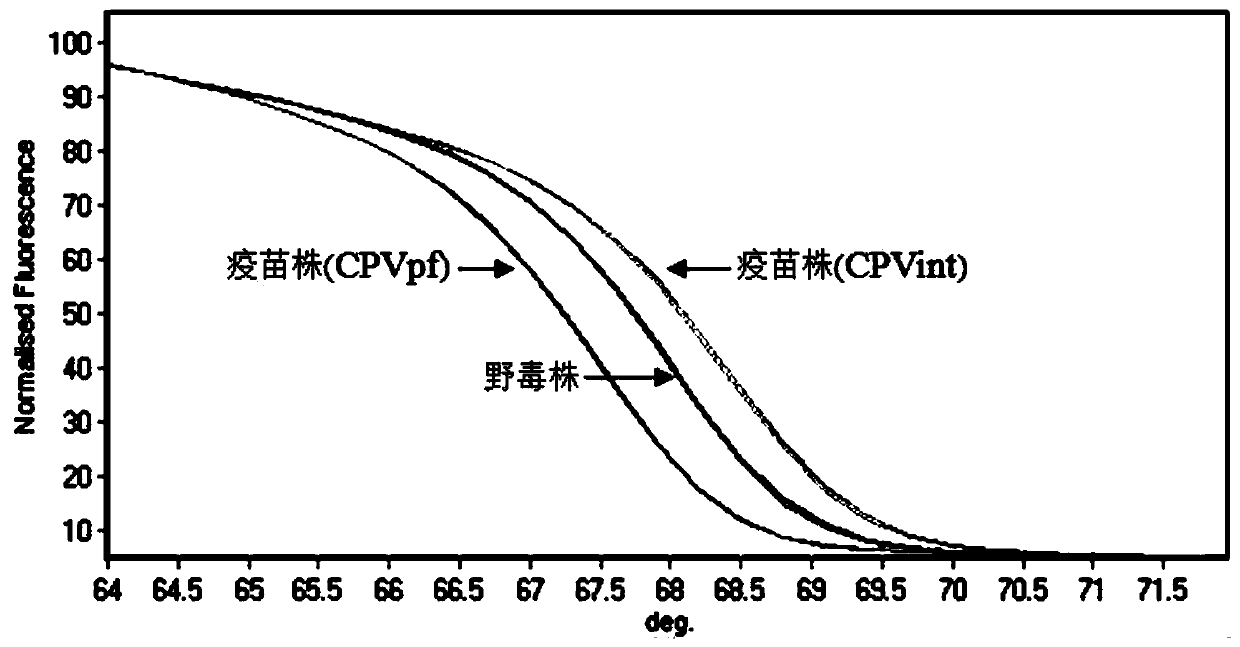
PCR-HRM primer and method for quickly distinguishing canine parvoviruses of different genotypes
ActiveCN104004857AQuick analysisEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationHigh fluxCanine parvovirus
The invention discloses a PCR-HRM primer and method for quickly distinguishing canine parvoviruses of different genotypes. The method comprises the following steps: extracting virus DNA (deoxyribonucleic acid) from a sample; by taking the virus DNA as a template, using a designed specific primer pair and fluorescent saturated dye to perform amplification reaction, thus obtaining an amplification product; and finally, performing HRM analysis on the amplification product, thus determining the genotype of a canine parvovirus. According to the invention, the method is simple to operate, only the fluorescent saturated dye needs to be added before PCR reaction, and the method is high in detection speed and high in flux; the whole operation process only costs 3 hours, and cell culture of viruses is not needed, thus greatly shortening the time required for typing; the expense is low, specific probes are not needed, and the cost of the saturated dye for each sample is 1.6RMB; and the accuracy is high, the specificity and repetitiveness are favorable, and analysis can be accurately and quickly performed at high flux, thereby ensuring that the invention is beneficial to popularization and application in clinical practice.
Owner:广东宠健生物科技有限公司 +1
RT-PCR-HRM or PCR-HRM primer, reagent and method for fast distinguishing four kinds of serotype dengue viruses
ActiveCN105779654AEasy to identifyEasy to operateMicrobiological testing/measurementMicroorganism based processesSerotypeGenome
The invention discloses an RT-PCR-HRM or PCR-HRM primer, reagent and method for fast distinguishing four kinds of serotype dengue viruses. According to the genomic sequence alignment result of the four kinds of serotype dengue viruses, three primers are designed, RNA is adopted as a template, an LC green dye is added for carrying out a one-step RT-PCR, or a built plasmid containing a dengue virus 280bp basic group is adopted as a template and the LC green dye is added for carrying out PCR amplification, the amplification product is subjected to HRM analysis, and the four kinds of serotype dengue viruses can be obviously distinguished through an HRM standard curve and a peak type figure. The method is easy to operate, the reagent is low in cost, high in detecting sensitivity and reliable in parting result, and the method is an innovative method for detecting and parting the four kinds of serotype dengue viruses.
Owner:GUANGDONG LAB ANIMALS MONITORING INST +1
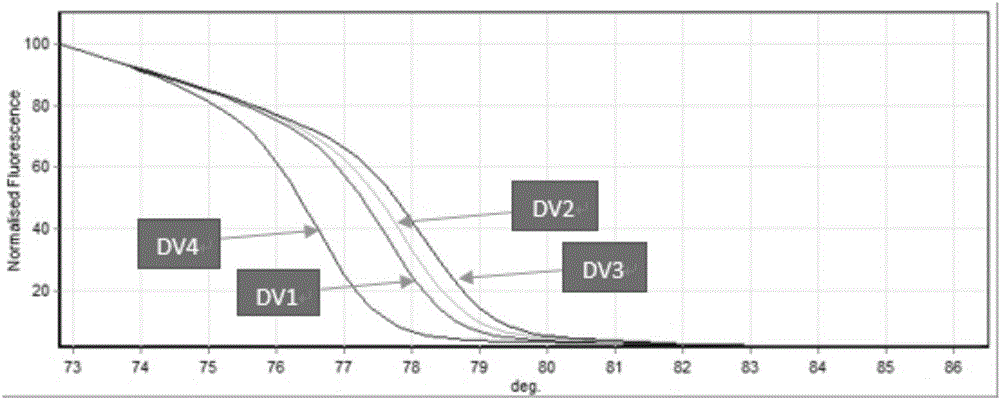
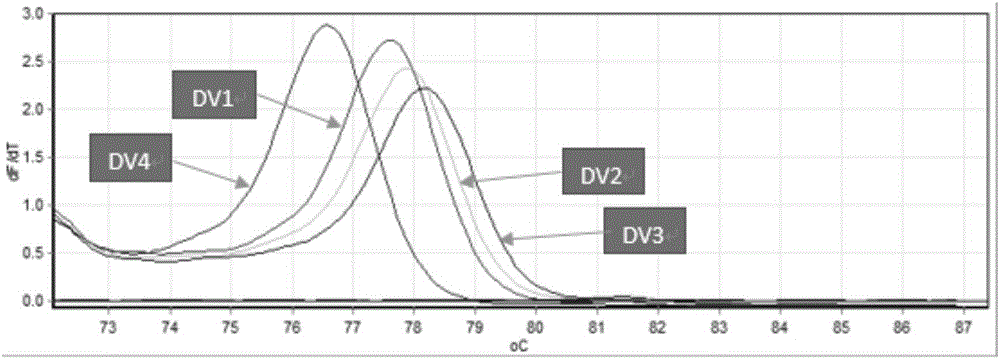
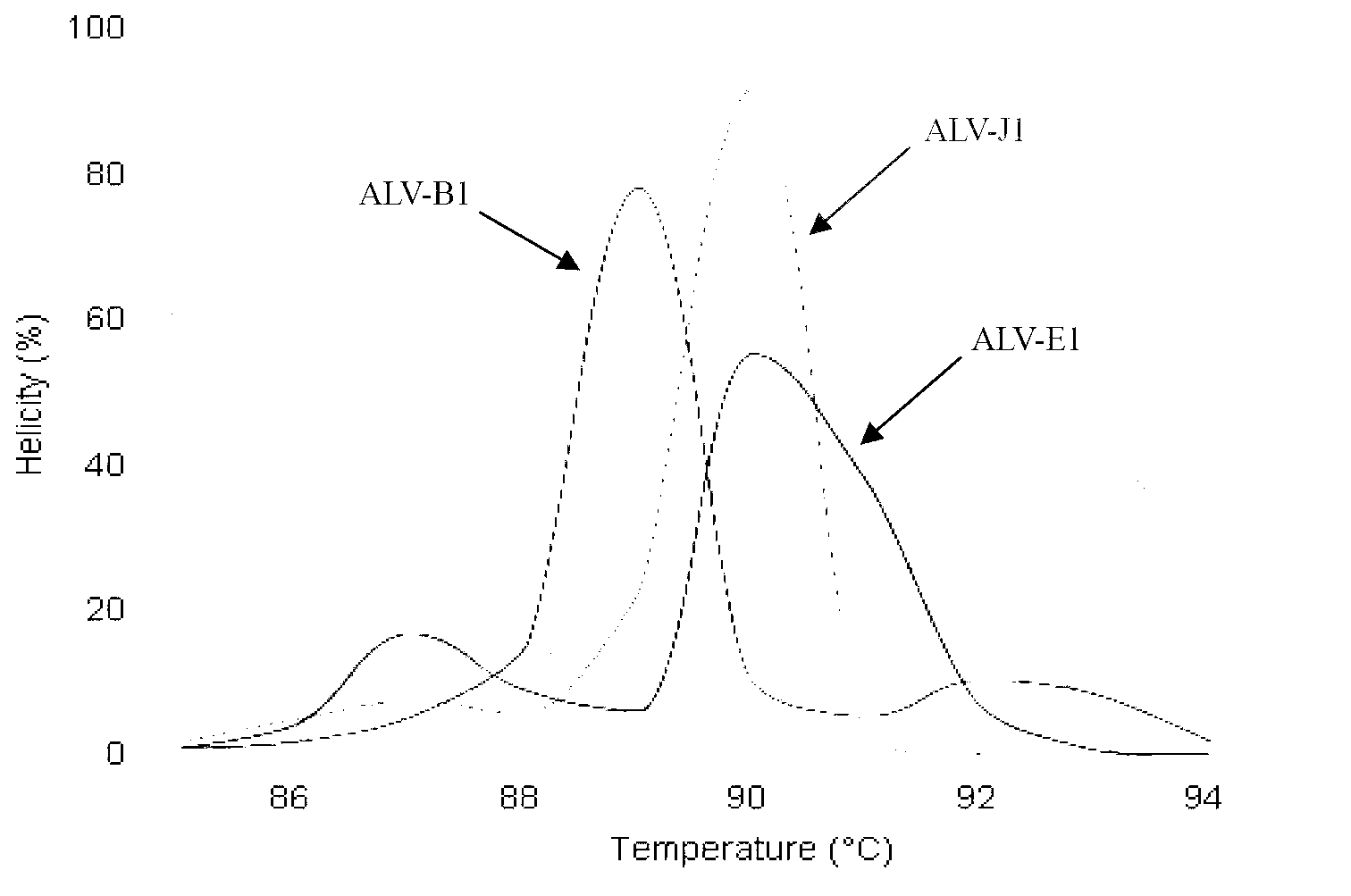
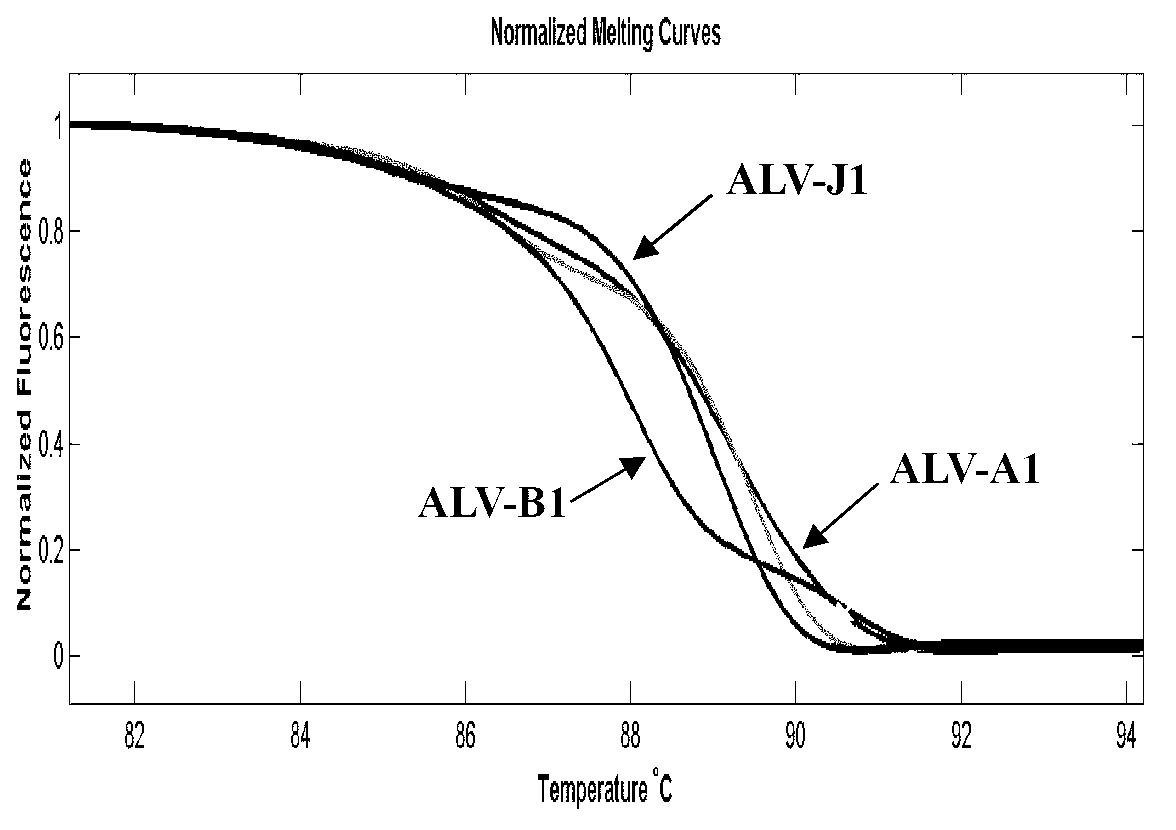
Establishment of PCR-HRM analysis method for rapid differential diagnosis of different serotypes of avian leukemia viruses
ActiveCN103074447AGood amplification effectReduce time to differential typingMicrobiological testing/measurementDNA/RNA fragmentationLeucosisSerotype
The invention discloses establishment of a PCR-HRM analysis method for rapid differential diagnosis of different serotypes of avian leukemia viruses. According to the invention, universal primers are good in degeneracy, have very good amplification performance to the A, B, C, D and E subtypes of avian leukemia viruses, is conductive to improving the efficiency of PCR (polymerase chain reaction), and reduces the time of identification and typing of viruses; and specific primers provided by the invention are good in specificity, and the J subtype can be amplified in a specific way. HRM analysis is performed on the amplification product obtained by using the primers, so that the typing of avian leukemia viruses can be performed accurately and rapidly in a high-throughput way by being compared with the standard HRM curve of a known subtype, particularly the endogenous type of avian leukemia viruses and the exogenous type of avian leukemia viruses can be rapidly and accurately distinguished.
Owner:INST OF ANIMAL HEALTH GUANGDONG ACADEMY OF AGRI SCI
SSR (Simple Sequence Repeat) core primer group and method thereof for identifying tea variety
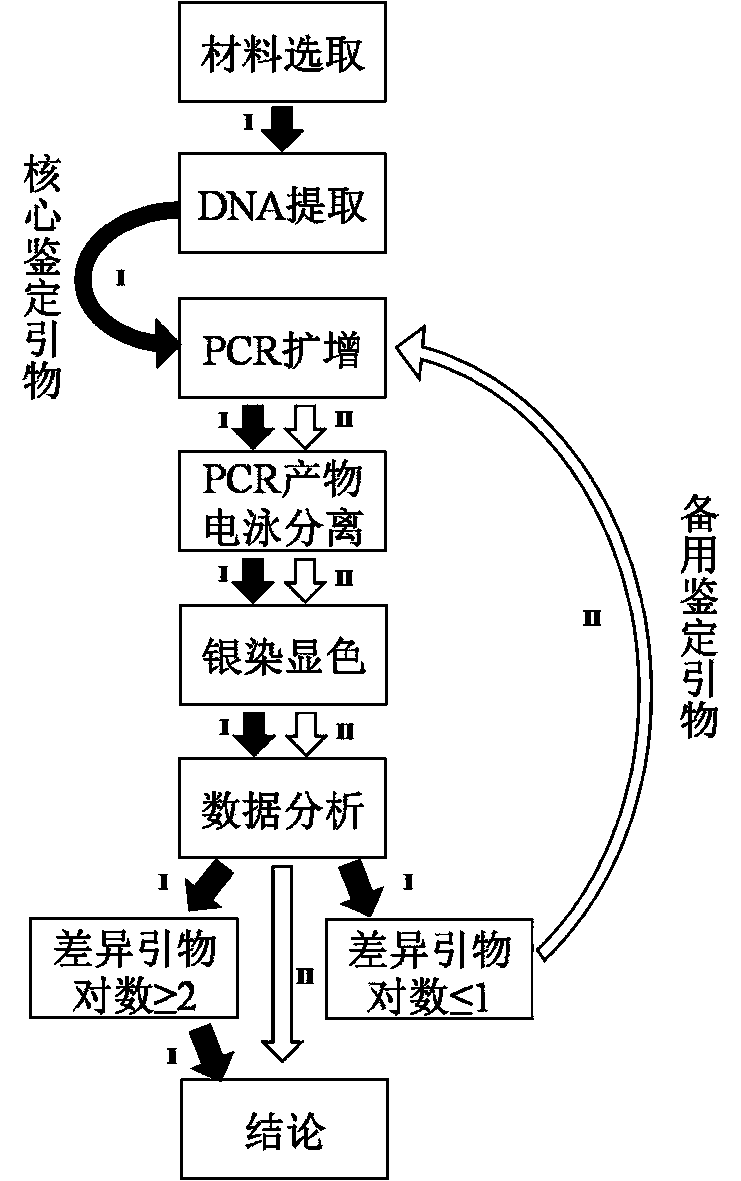
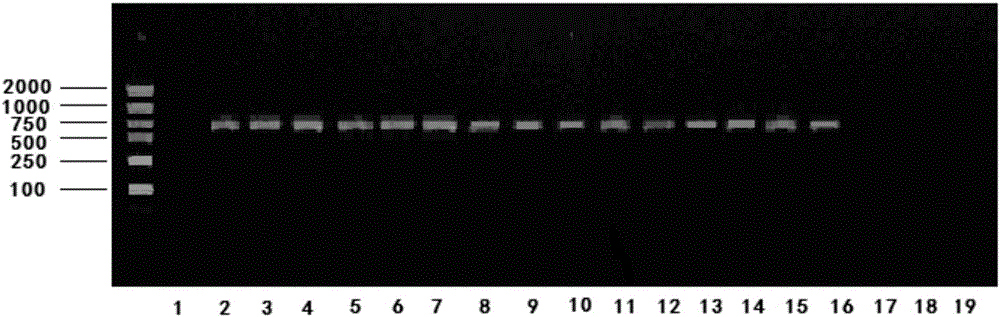
InactiveCN104004756AImprove identification efficiencyAvoid identification errorsMicrobiological testing/measurementDNA/RNA fragmentationElectrophoresisDNA extraction
The invention relates to an SSR (Simple Sequence Repeat) core primer group and a method thereof for identifying a tea variety, and belongs to the technical field of biology. The method is characterized by comprising the following steps: (1) material selection; (2) genome DNA extraction; (3) PCR (Polymerase Chain Reaction) amplification of a target product; (4) electrophoretic separation of a PCR product; (5) silver staining development; (6) data analysis; and (7) result judgment. The SSR core primer group and the method thereof for identifying the tea variety, which are disclosed by the invention, have the characteristics of easiness and convenience for operation, accuracy in result, high repeatability, high identification capacity, low cost and the like and are suitable for identifying the tea variety.
Owner:TEA RES INST CHINESE ACAD OF AGRI SCI
Method for identifying donkey skin from counterfeit species by applying mitochondria COI (cytochrome oxidase subunit I) sequence segments and specific primer for amplifying segments
ActiveCN105112417AGood amplification effectEasy to operateMicrobiological testing/measurementDNA preparationBiologySequence assembly
The invention discloses a method for identifying donkey skin from counterfeit species by applying mitochondria COI (cytochrome oxidase subunit I) sequence segments and a specific primer for amplifying the segments, belonging to the technical field of identification of Chinese medicine. According to the method disclosed by the invention, COI sequences of the donkey skin and the counterfeit species are amplified by using an autonomously designed primer, the donkey skin is distinguished from pigskin or cowhide by judging whether a PCR (Polymerase Chain Reaction) product exists in an amplified band in gel electrophoresis or not, and mitochondria COI sequences of the donkey skin from multiple sources and common counterfeit species are subjected to sequence assembly and compared, and then the donkey skin and counterfeit species thereof are intuitively identified through a K2P distance method. The method disclosed by the invention is convenient and rapid and high in accuracy and has the capability of well ensuring the quality of donkey-hide gelatin and donkey skin medicinal materials.
Owner:SHANDONG UNIV OF TRADITIONAL CHINESE MEDICINE
Primer, kit and method for identifying dove sex
ActiveCN105925690AStrong specificityHigh resolutionMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionMicrobiology
The invention discloses a primer, a kit and a method for identifying dove sex. The primer has sequences as shown in SEQ ID NO: 1 and SEQ ID NO: 2; the primer has the advantages of being good in specificity, high in resolution ratio, good in amplification effect and the like; and the primer, without enzyme digestion, can directly conduct agarose gel electrophoresis. Moreover, two specific products, compared with a specific product, are higher in recognition degree, so that an amplification result can be recognized at a glance; the invention also provides a method for identifying the dove sex by virtue of the primer; the method, under the precondition of conducting specific amplification by virtue of the primer, can also reduce time cost and economic cost of identifying the dove sex by simplifying a sample DNA acquisition process; and the method, which identifies the dove sex by virtue of the primer, is broad in market utilization value.
Owner:深圳市天翔达鸽业有限公司
Kit for instantly detecting vaginitis pathogen gene
InactiveCN105420360ASimple compositionLow costMicrobiological testing/measurementFluorescencePhosphate
The invention discloses a kit for instantly detecting a vaginitis pathogen gene. The kit is used for amplification and detection of a gene at constant temperature. The kit can perform rapid fluorescence detection through matching between an isothermal amplification reaction reagent and a two-color fluorescent labeled probe. The isothermal amplification system of the kit really achieves gene amplification reaction at the same temperature, does not depend on ATP, creatine phosphate and other high-energy energy molecules, is simpler in composition, lower in cost and more convenient to use compared with an existing isothermal amplification system. The isothermal amplification system is suitable for amplification of complex templates, is high and stable in amplification speed, can complete amplification reaction within 15 minutes, high in amplification sensitivity and accuracy, does not produce mismatch and is good in amplification effect.
Owner:GUANGZHOU HEAS BIOTECH CO LTD
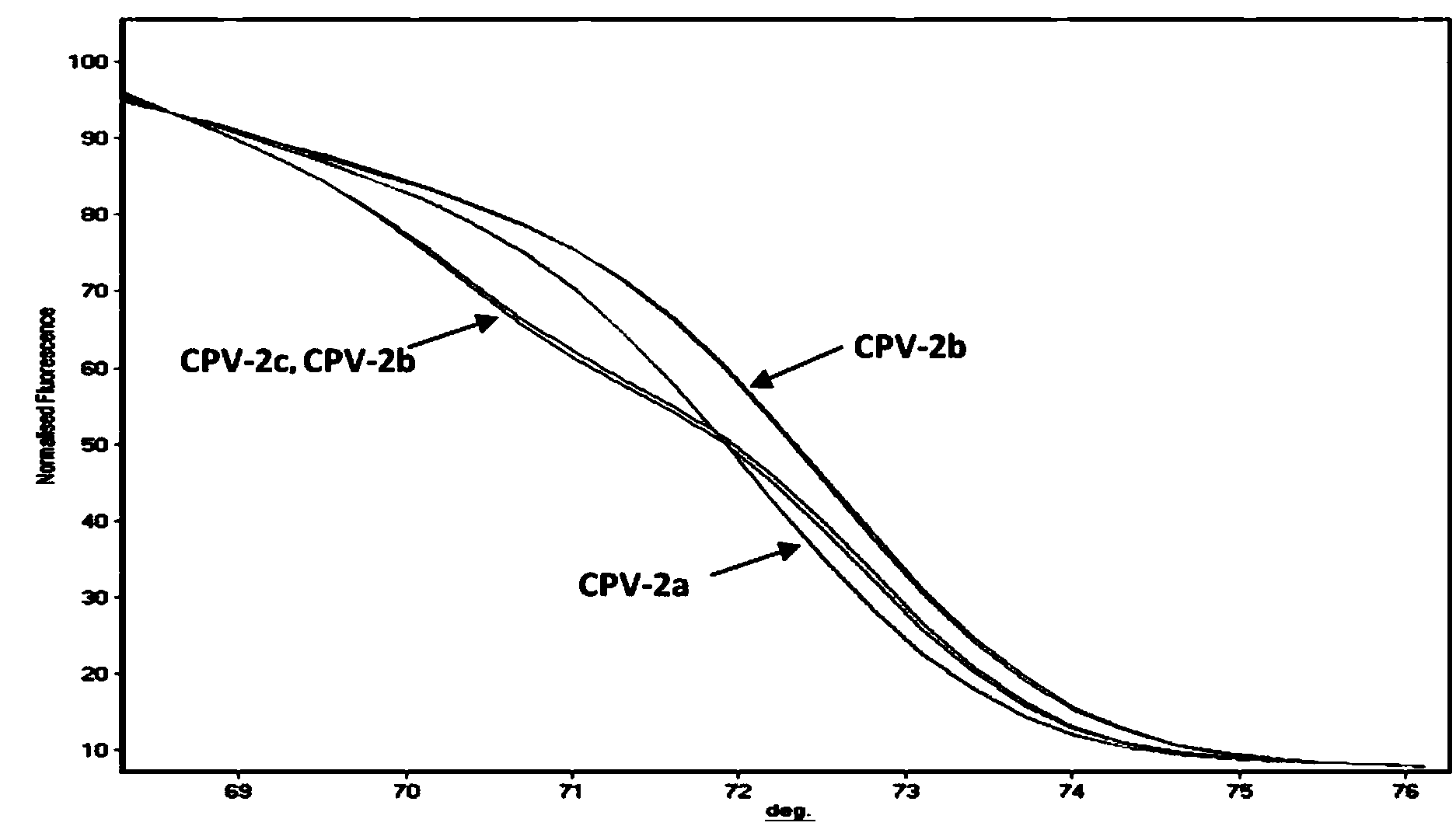
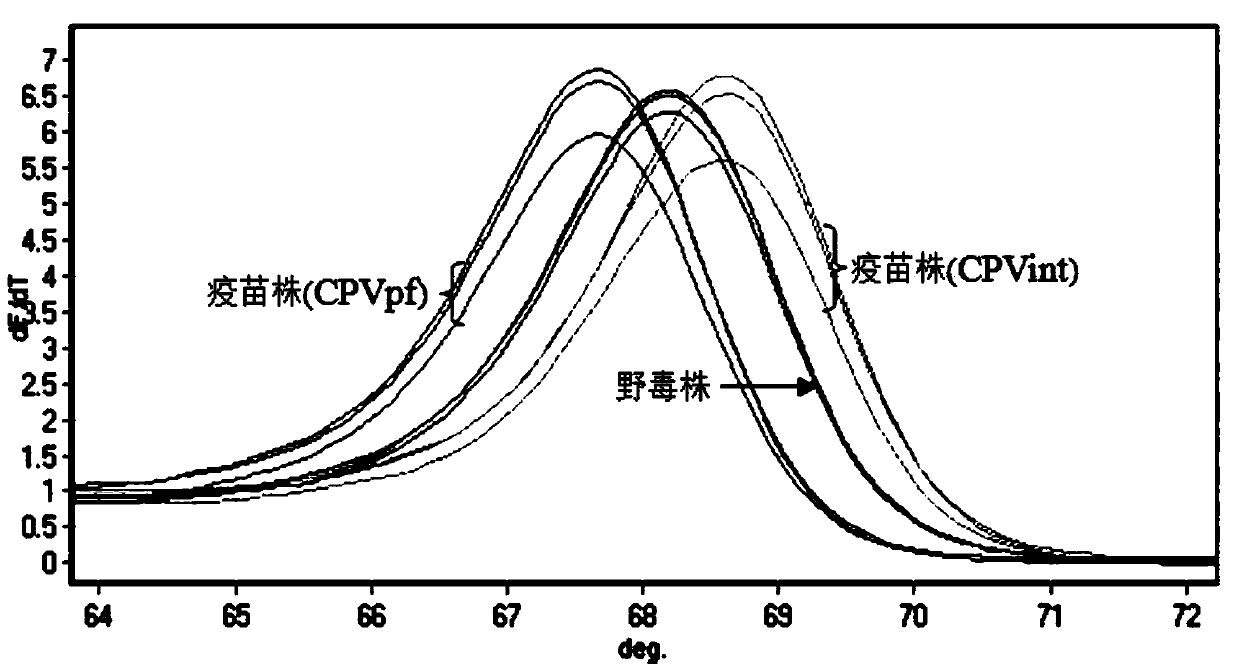
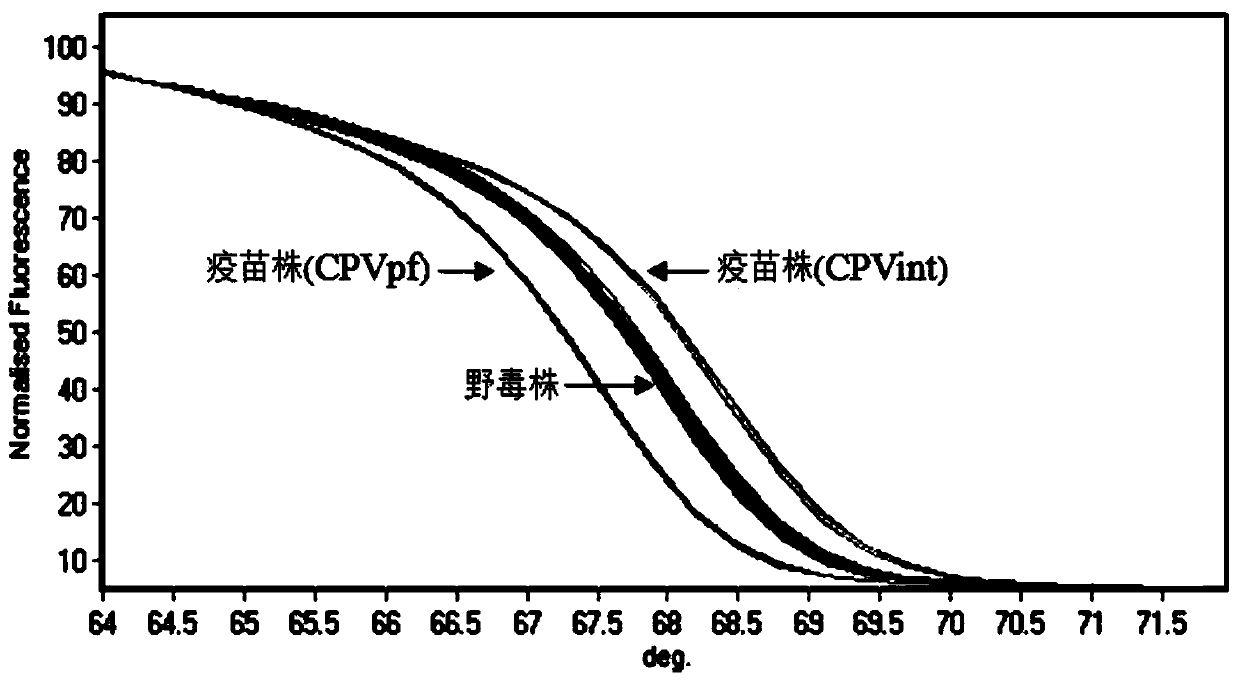
PCR-HRM primer and method for rapidly distinguishing wild strain and vaccine strain of canine parvovirus
ActiveCN104195265AQuick analysisEasy to operateMicrobiological testing/measurementMicroorganism based processesCanine parvovirusTGE VACCINE
The invention discloses a PCR-HRM primer and a method for rapidly distinguishing a wild strain and a vaccine strain of canine parvovirus. The method comprises the following steps of: extracting a virus DNA from a sample as a template, carrying out PCR amplification by utilizing designed two pairs of specific primers and fluorescent saturated dye, carrying out HRM analysis on the detected sample respectively with a wild strain standard sample and a vaccine strain standard sample as contrast, and determining the type of the canine parvovirus. With the adoption of the method, the operation is simple, and only is the fluorescent saturated dye added before the PCR; the detection speed is high, the flux is high, the whole operation process only needs 3 hours, and cell culture of viruses is not needed, and thus the time needed by distinguishing detection is greatly shortened; the cost is low, a specific probe is not needed, and the fluorescent saturated dye is cheap and easily obtained; as the accuracy is high, the specificity and the repeatability are good, and analysis can be accurately and rapidly carried out with high flux, the method is beneficial to popularizing and applying in clinical practices.
Owner:INST OF ANIMAL HEALTH GUANGDONG ACADEMY OF AGRI SCI
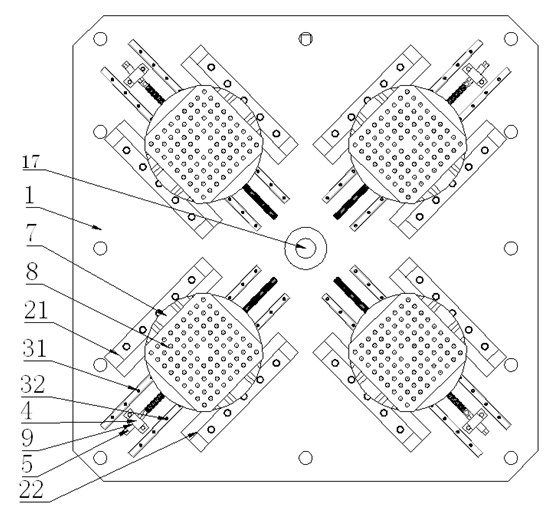
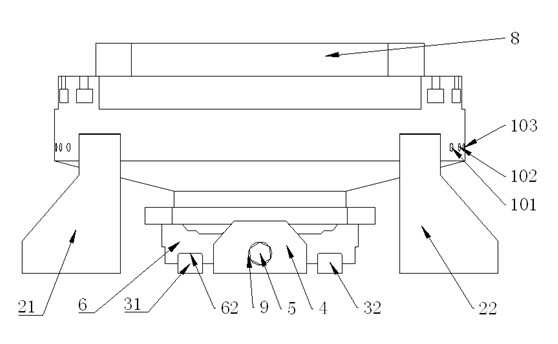
Vibration table fixture capable of accurately adjusting center of gravity
InactiveCN102156034AHigh positioning accuracyHigh precisionVibration testingStructural engineeringTwo degrees of freedom
The invention discloses a vibration table fixture capable of accurately adjusting the center of gravity, comprising a vibration table top and four subsidiary fixtures in same structures, wherein each subsidiary fixture comprises two guiding supporting plates, two sliding bars, a supporting block, screws, a sliding block, a supporting disc and a base; the four subsidiary fixtures are symmetrically distributed by using a center hole of the vibration table top as the center; the center lines of the screws of every two adjacent subsidiary fixtures form an included angel of 90 degrees; and the center lines of the screws of every two opposite subsidiary fixtures coincide with each other. By using the invention, the base can be adjusted in two degrees of freedom relative to the vibration table top, and the coincided centers of gravity of both a large test piece arranged on the base and the vibration table fixture can coincide with the axis of the vibration table when the large test piece is under the vibration test. The invention can ensure that the center of gravity of the test piece passes by the center of the vibration table top to reduce the influence of the overturning moment and can realize the rigid connection of the test piece and the vibration table top or the vibration table fixture.
Owner:ZHEJIANG UNIV
Application of DNA target sequence of schistosoma japonicum retrotransposon in schistosomiasis diagnosis
ActiveCN102010861AElucidating genetic differencesGood amplification effectMicrobiological testing/measurementEnzymesNucleotideA-DNA
The invention discloses application of a DNA target sequence of schistosoma japonicum retrotransposon in schistosomiasis diagnosis and particularly provides retrotransposon shown by any one nucleotide sequence selected from SEQ ID NO:1-25 and can be used as a detection marker for schistosomiasis. The invention also provides a pair of primers for specifically amplifying the retrotransposon sequence.
Owner:CHINESE NAT HUMAN GENOME CENT AT SHANGHAI +1
Primer, method and kit for constructing gene mutation sequencing library
ActiveCN104946639AGood amplification effectMultiplex PCR amplification effect is goodMicrobiological testing/measurementLibrary creationMicrobiologyA-DNA
The invention relates to a PCR primer involving 47 genes and a tag PCR primer. The tag PCR primer is composed of the PCR primer and a DNA tag sequence connected to the 5' end of at least one primer in the PCR primer pair, wherein the DNA tag sequence is selected from SEQ ID NO: 107 to SEQ ID NO: 126. The invention further relates to a method and a kit for constructing a sequencing library by applying the DNA tag of the tag PCR primer. The coincidence rates of the detection method provided by the invention and the sequencing method is up to 100%, the provided amplification primer and the DNA tag form a detection product with a specificity, a sensitivity and a repeatability which are optimized and balanced, somatic mutation and mononucleotide polymorphism can be detected in parallel, and a plurality of base sequences of 1-20 sample sources can be accurately differentiated by one-time detection.
Owner:广州益善医学检验所有限公司
Quick batch extraction method for cotton DNA (Deoxyribonucleic Acid) suitable for PCR (Polymerase Chain Reaction)
The invention discloses a quick batch extraction method for cotton DNA (Deoxyribonucleic Acid) suitable for PCR (Polymerase Chain Reaction). The method comprises the following steps: filling the top end of the hypocotyledonary axis of a cotton seedling in a hole of a PCR plate; adding NaOH liquor; crushing cells of a sample in a boiling water bath; taking the sample out and adding Tris-Hcl liquor; then putting the sample liquor in a boiling water bath; taking the sample liquor out and cooling; and diluting the sample liquor for 10-30 times by ddH2O to obtain a PCR template of cotton DNA. The banding pattern of the cotton DNA obtained by adopting the quick batch extraction method is highly consistent with that of the DNA extracted by a conventional CTAB (Cetyltrimethyl Ammonium Bromide) method through detection of an SSR (Simple Sequence Repeat) molecular marker. The method disclosed by the invention is simple and quick, fussy procedures of the CTAB method and a SDS (Sodium Dodecyl Sulfate) method are avoided, and time and labor are saved. The method disclosed by the invention is suitable for projects such as DNA fingerprint purity detection of crossbred species of cotton in batches and cotton breeding improvement needing to extract DNA in batches for detection.
Owner:HEFEI FENGLE SEED
Primer pair, kit, method for identifying true and false of cordycepssinensis and application thereof
PendingCN109825624AWide range of identificationStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationBiology
Owner:DONGGUAN HEC CORDYCEPS R&D CO LTD
Universal test primer for viruses in tobamovirus and method for universal test primer
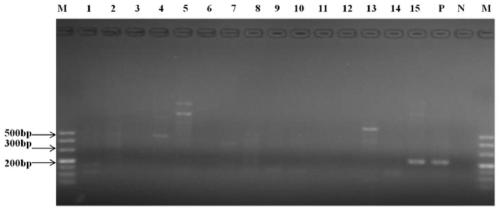
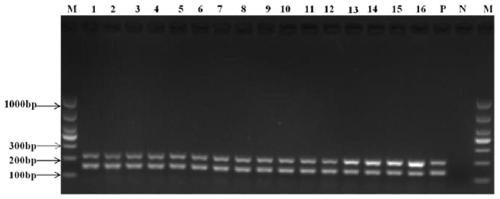
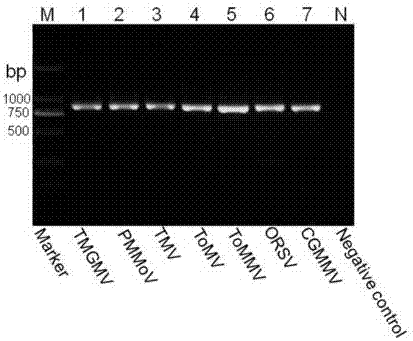
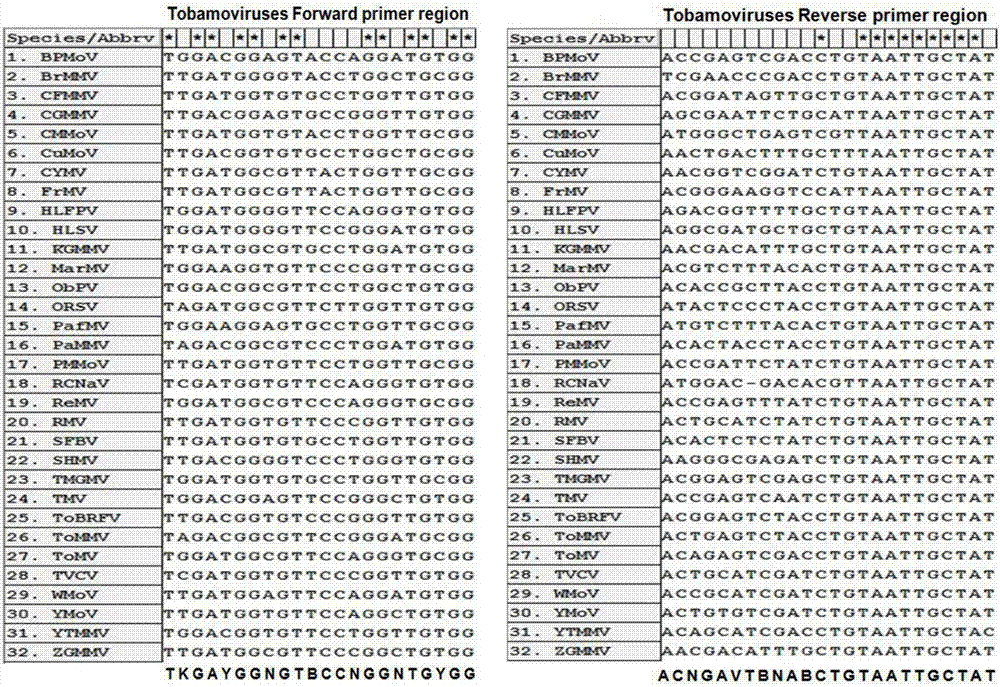
InactiveCN107058611AGood amplification effectImprove detection efficiencyMicrobiological testing/measurementMicroorganism based processesForward primerVirus
The invention belongs to the technical field of molecular biology and virology and particularly relates to a universal test primer for viruses in a tobamovirus and a method for the universal test primer. Primer pair sequences of the universal test primer for the viruses in the tobamovirus are a forward primer TobamodF:5'-TKGAYGGNGTBCCNGGNTGYGG-3', and a reverse primer TobamodR:5'-ACNGAVTBNABCTGTAATTGCTAT-3. The test primer is designed on the basis of 32 conserved region sequences of a virus genome in the tobamovirus, amplification effect verification is carried out on the pair of primers by using seven viruses in the tobamovirus, and the amplification effects all are relatively good, thereby building universal test primer and method capable of testing at least seven viruses in the tobamovirus. The universal test primer has the characteristics of being high in testing efficiency, high in sensitivity and low in cost.
Owner:YUNNAN AGRICULTURAL UNIVERSITY
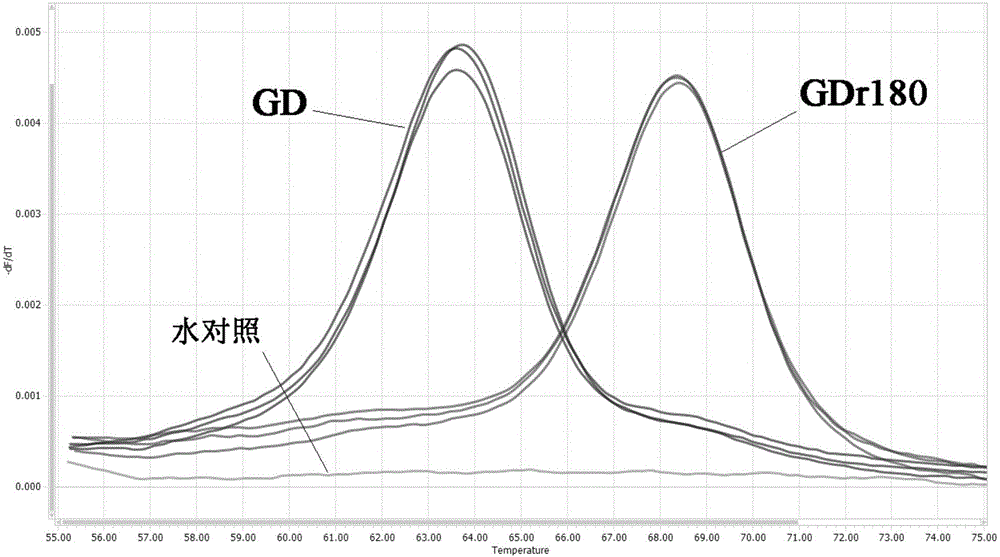
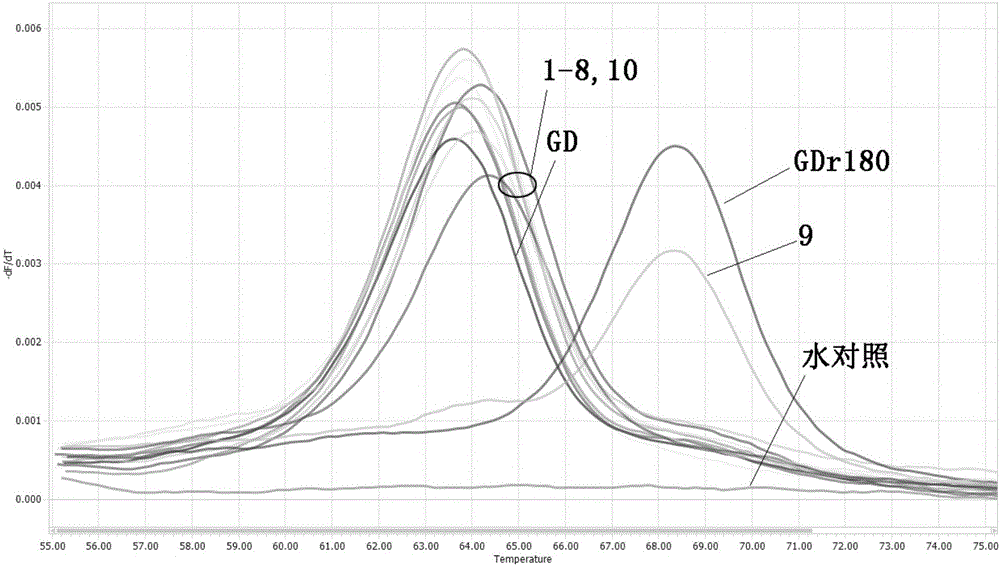
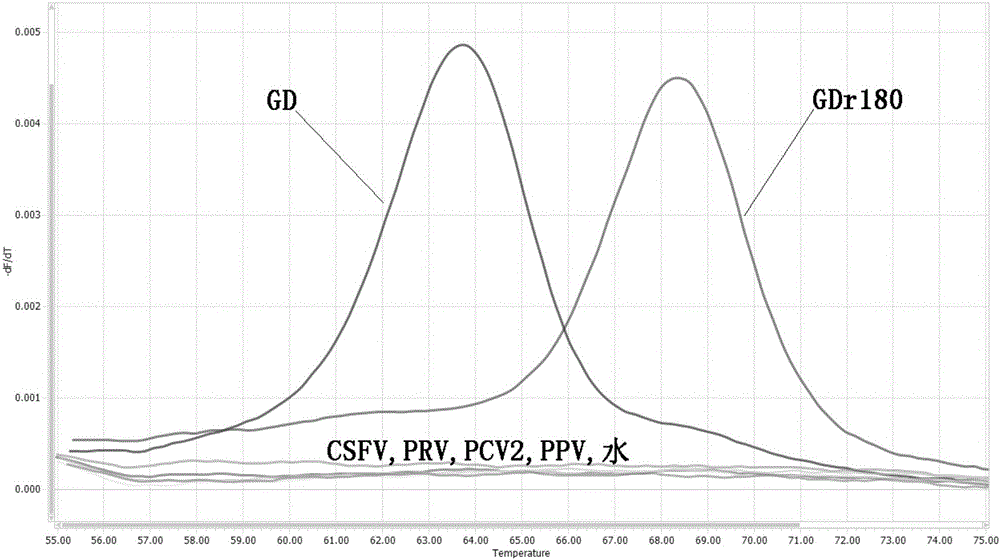
Primers, probe and method for rapidly distinguishing HP-PRRS (High pathogenic porcine reproductive and respiratory syndrome) vaccine strain GDr180 from HP-PRRS wild strain
ActiveCN105907890AEasy to operateDetection speedMicrobiological testing/measurementDNA/RNA fragmentationWild strainBiology
The invention discloses primers, probe and method for rapidly distinguishing an HP-PRRS (High pathogenic porcine reproductive and respiratory syndrome) vaccine strain GDr180 from an HP-PRRS wild strain. The method combines a real-time PCR (Polymerase Chain Reaction) technology with an MCA (Melting Curve Analysis) technology, and according to a difference of Tm values of a melting curve, the strain GDr180 and the wild strain are identified; the primers, the probe and the method are simple to operate, i.e. only the probe needs to be added before a PCR reaction; a detection speed is high and flux is high, i.e. the entire operating process only needs 3 hours, and time required for parting is greatly shortened; cost is relatively low, i.e. the identifying and detecting aims can be fulfilled only by one common probe; accuracy is high, specificity is good, repeatability is good, analysis can be accurately and rapidly carried out under the high flux, and the primers, the probe and the method are beneficial to popularization and application in clinical practice.
Owner:INST OF ANIMAL HEALTH GUANGDONG ACADEMY OF AGRI SCI
Triple fluorogenic quantitative PCR detection primer, kit and method of DTMUV, EDSV and H9 subtype AIV
ActiveCN104894292AGood amplification effectGood repeatabilityMicrobiological testing/measurementMicroorganism based processesEscherichia coliDisease
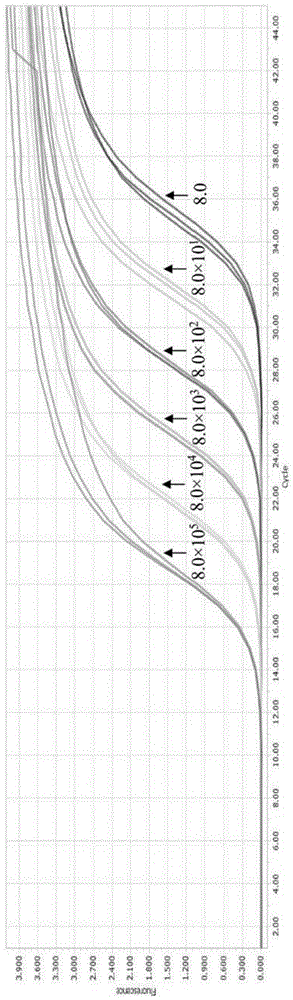
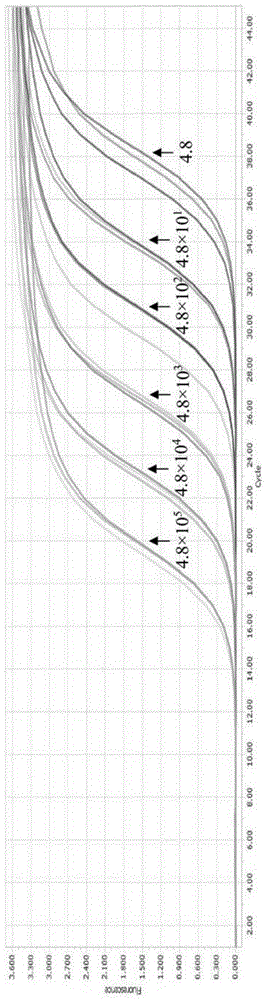
The invention discloses a triple fluorogenic quantitative PCR detection primer, kit and method of the duck tembusu virus disease, the egg drop syndrome virus and the H9 subtype avian influenza virus. The detection primer for the duck tembusu virus disease, the egg drop syndrome virus and the H9 subtype avian influenza virus is shown in SEQ ID NO.1-6. The detection primer and kit can detect the EDSV, the H9 AIV and the DTMUV at the same time. No cross reaction with duck plague virus, goose parovovirus, muscovy duck parvovirus and escherichia coli genomic DNA and duck hepatitis A virus 1, duck hepatitis A virus 3, muscovy duck reovirus and newcastle disease virus RNA exists. The lower limits of detection for EDSV nucleic acid, H9 AIV nucleic acid and DTMUV nucleic acid are 8.0 copies, 4.8 copies and 1.3 copies respectively. The detection primer, kit and method can be used for qualitative and quantitative detection for the EDSV, the H9 AIV and the DTMUV.
Owner:INST OF ANIMAL HEALTH GUANGDONG ACADEMY OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com