SSR (Simple Sequence Repeat) core primer group and method thereof for identifying tea variety
A core primer and variety technology, applied in the biological field, can solve the problems of low efficiency, poor identification result accuracy, time-consuming and labor-intensive problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
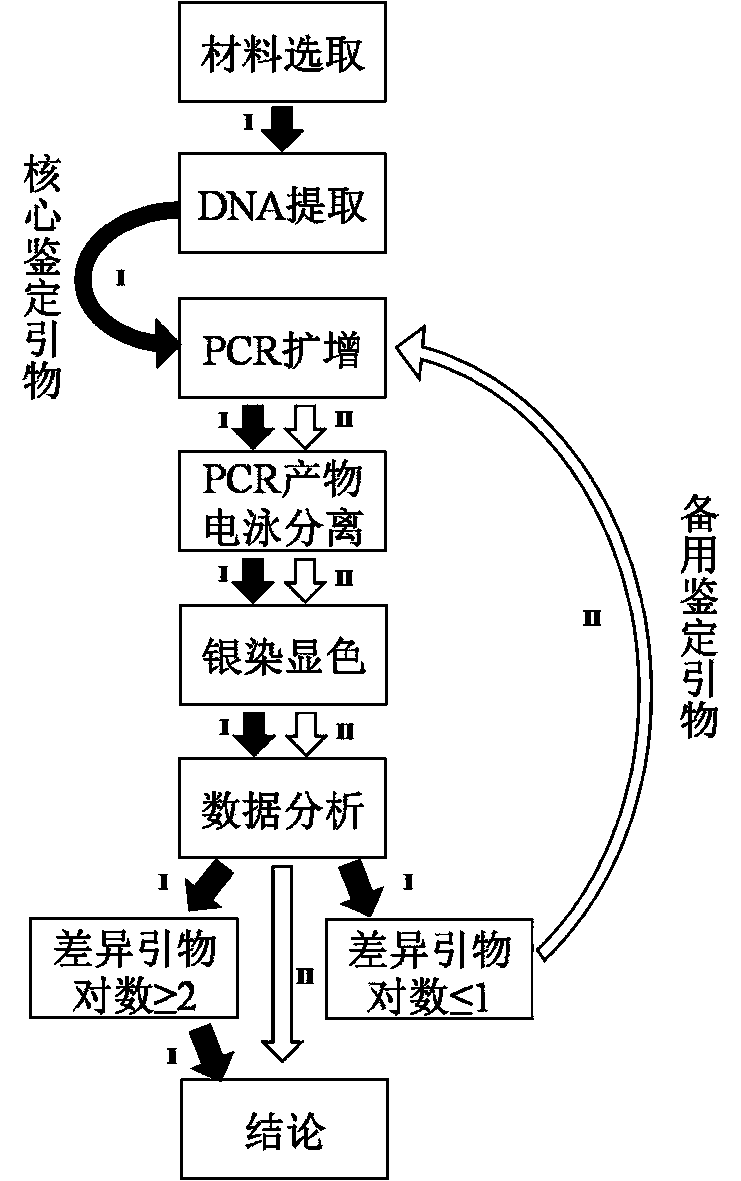
Examples
Embodiment 1
[0059] 1) Use and preparation of main reagents:
[0060] ① Genomic DNA extraction kit: a quick plant genome extraction kit produced by Beijing Tiangen Biochemical Technology Co., Ltd.;
[0061] ②Reagents required for PCR reaction: use rTaq enzyme produced by Dalian Bao Biological Engineering Co., Ltd., including 50 μL 5U / μL rTaq enzyme, 1 mL 10×Buffer, 800 μL dNTP Mixture and 1 mL Loading Buffer;
[0062] ③1L 5×TBE: 54g Tris, 27.5g boric acid, 20mL 0.5M EDTA (pH 8.0);
[0063] ④500mL 30% acrylamide: 145g acrylamide and 5g N, N-dimethylene bisacrylamide, add ddH 2 O is fixed to 500mL;
[0064] ⑤50mL 10% ammonium persulfate: 5g ammonium persulfate, add ddH 2 O is fixed to 50mL;
[0065] ⑥1L fixative: 100ml ethanol solution, 5mL glacial acetic acid, add distilled water to make up to 1L;
[0066] ⑦1L staining solution: 2g AgNO 3 , add distilled water to make up to 1L;
[0067] ⑧1L chromogenic solution: Prepared with distilled water, 15g NaOH solution and added 5mL 37% forma...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com