PCR-HRM primer and method for quickly distinguishing canine parvoviruses of different genotypes
A canine parvovirus, genotype technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of complex operation, limited application, high cost, and achieve good repeatability, The effect of good degeneracy and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
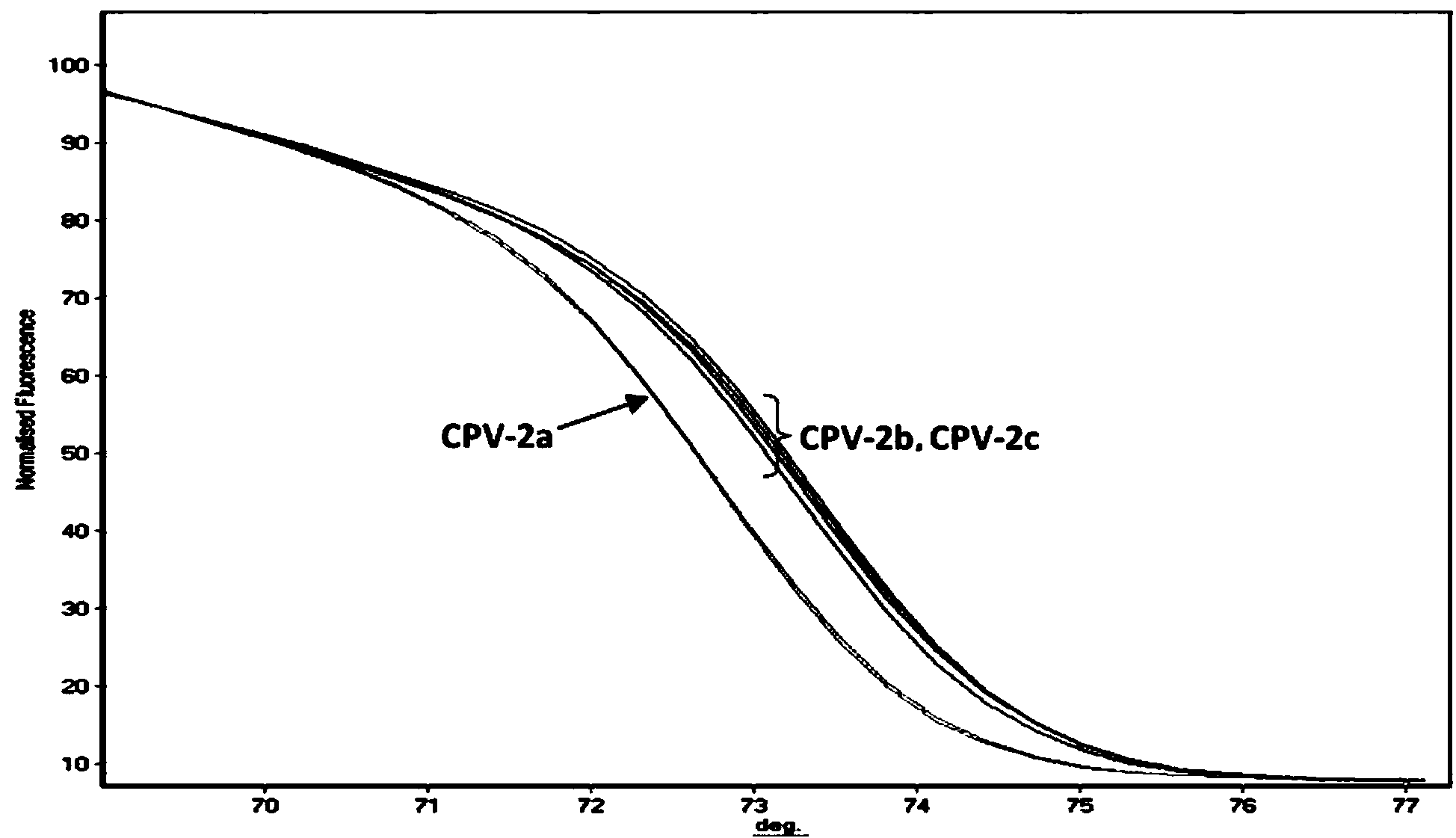
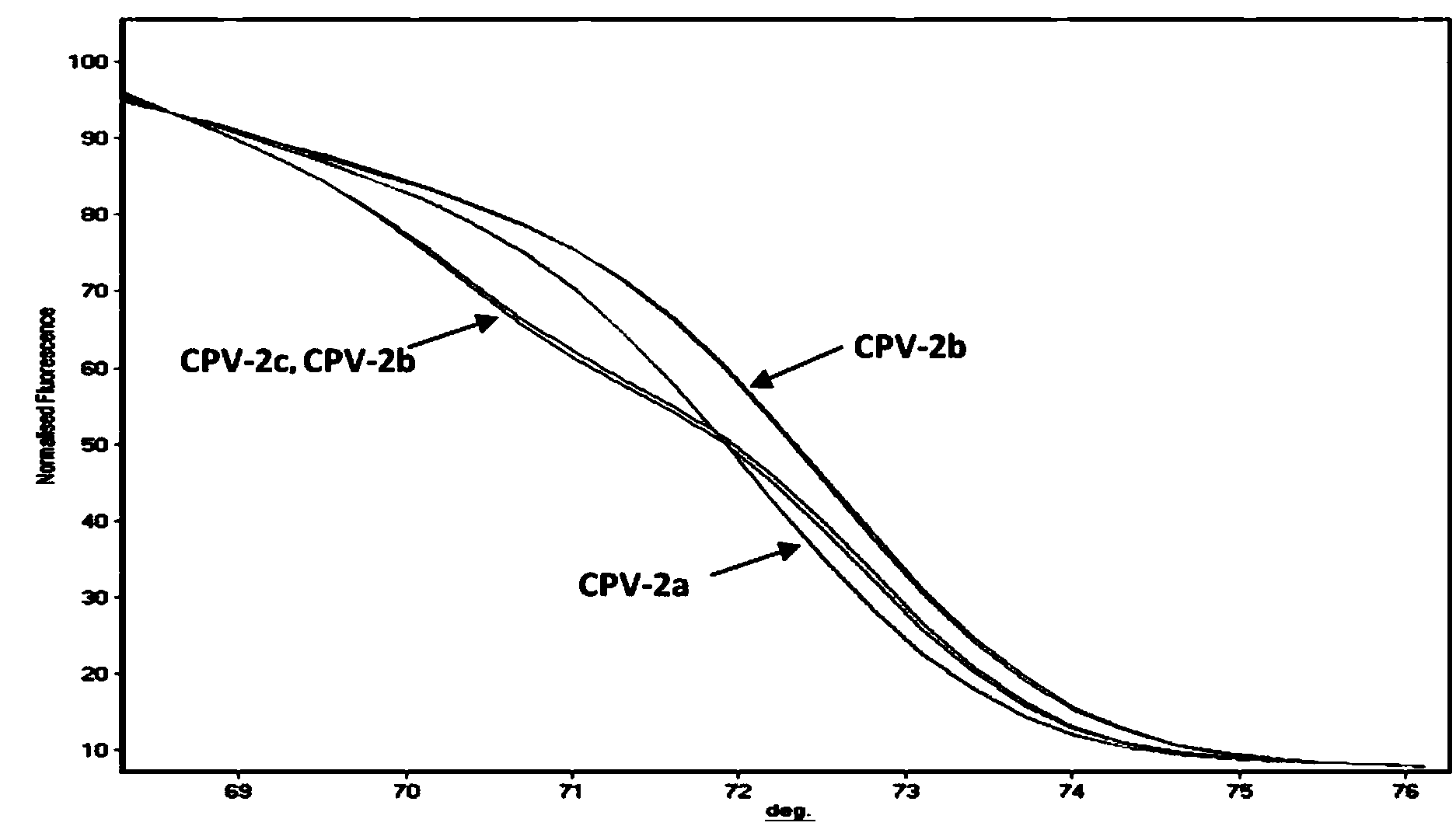
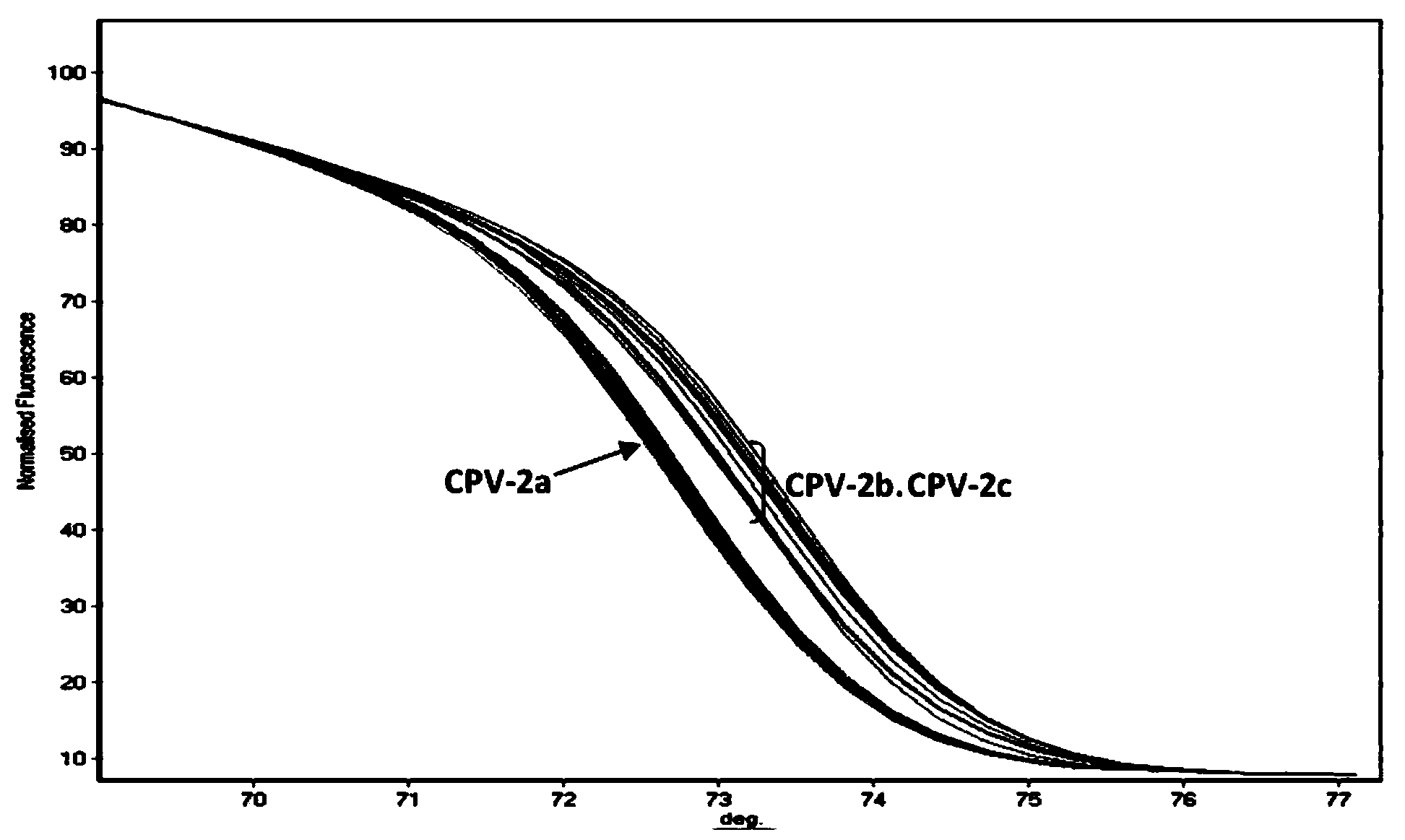
[0061] (1) PCR-HRM primers
[0062] After screening a large number of designed primers, it was found that the base sequences of the primers, SEQ ID NO: 1 and SEQ ID NO: 2, have the best effect on the PCR-HRM method for distinguishing different genotypes of canine parvovirus, and the base sequences are as follows Show.
[0063] P1 : 5'-CCAGAAGGAGATTGGATTCA-3' (SEQ ID NO: 1),
[0064] P2: 5'-TTAATGCAGTTAAAGGACCAT-3' (SEQ ID NO: 2).
[0065] Among them, the primer P1 is on the 4014-4033 position of the canine parvovirus VP2 gene, and the primer P2 is on the 4138-4158 position of the canine parvovirus VP2 gene (the accession number of this gene in Genbank is M38245).
[0066] (2) Construction of standard plasmid samples and PCR-HRM analysis
[0067] 1) Extraction of canine parvovirus DNA:
[0068] Canine parvovirus DNA in disease samples was extracted with MiniBEST Viral RNA / DNA Extraction Kit Ver.4.0. Disease samples can be whole blood, feces, rectal swabs and other samp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com