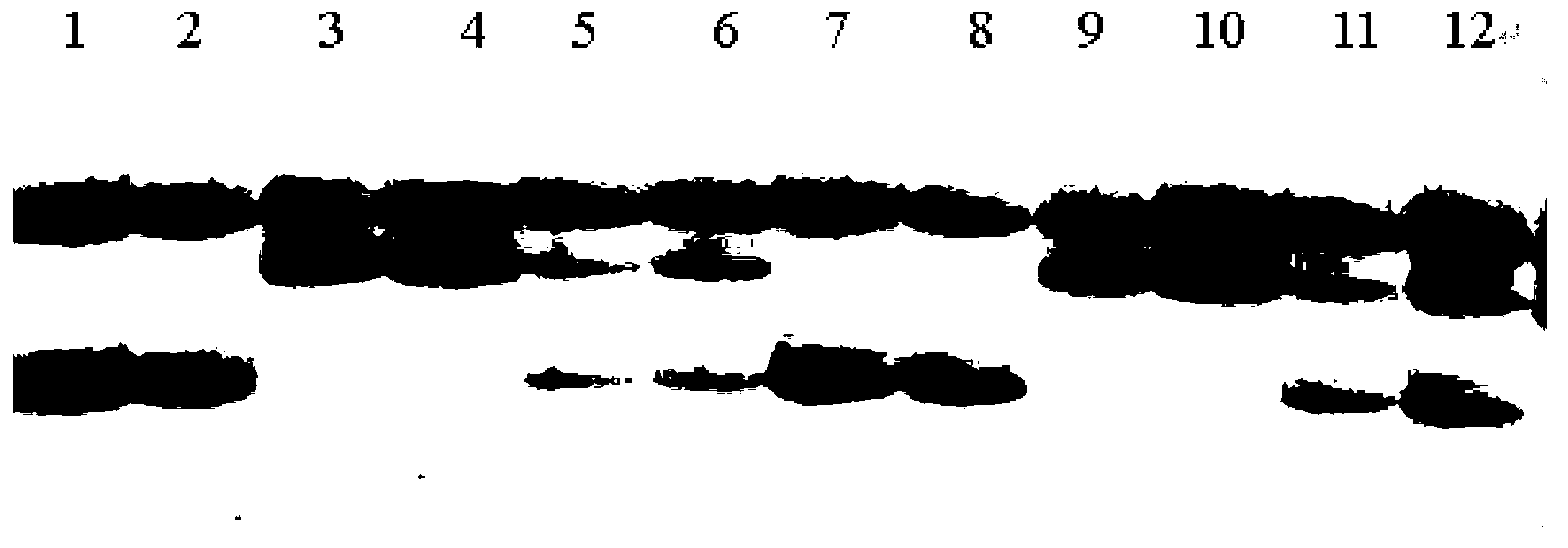Quick batch extraction method for cotton DNA (Deoxyribonucleic Acid) suitable for PCR (Polymerase Chain Reaction)
A cotton, batch technology, applied in the field of rapid batch extraction of cotton DNA, can solve the problems of increasing the complexity of cotton DNA extraction, affecting molecular biology operations, and unsatisfactory, achieving fast and easy waiting for cotyledons to obtain materials, and fast and easy DNA extraction time , the effect of consistent strip size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 (a method for rapid batch extraction of cotton DNA suitable for PCR in the present invention).
[0019] Take Guofeng Cotton 198 as an example: Take the parental and standard F1 generation of Guofeng Cotton 198, and germinate indoors on sandy soil. After 5-6 days, two cotyledons are unfolded, and the sample can be collected.
[0020] Follow the steps below:
[0021] 1) Take the 0.45-0.55cm of the above two plants of the female parent, male parent and F1 generation at the top of the hypocotyl of healthy cotton in the seedling stage, and place them in each well of a 96-well PCR plate (preferably on ice Operation), and add 40μL of 0.25mol / L NaOH solution to each well;
[0022] 2) Add step 1) the top of hypocotyl and the PCR plate with NaOH solution to each well, and boil it in boiling water in a water bath for 1-1.5min;
[0023] 3) Add 60μL of 0.17mol / L Tris-Hcl solution to each well of the PCR plate after step 2) boiled, and then place it in boiling water in a water bath ...
Embodiment 2
[0026] Example 2 (using the current improved CTAB cotton genomic DNA extraction method, as a comparative example).
[0027] ① The samples were collected by collecting cotyledons from the same plants of the female parent, male parent, and F1 generation of Guofeng Cotton 198 taken in Example 1, put them into 2.0 mL centrifuge tubes, and add 600 μL of freshly prepared extraction buffer , Grinding machine for grinding.
[0028] ②Centrifuge at 10000rpm for 10min (4℃), discard the supernatant.
[0029] ②Add 600 μL of 65 ℃ preheated lysis buffer to the precipitate, and use a copper wire or toothpick to stir and mix well. In a 65 ℃ water bath for 30 minutes, gently shake once every 10 minutes.
[0030] At the end of the water bath, add 600μL of chloroform: isoamyl alcohol (24:1) mixture and turn over 30 times until the lower solution turns black.
[0031] ⑤Centrifuge at 10000rpm for 10min (room temperature), transfer the supernatant to a 1.5mL centrifuge tube, add an equal volume of pre-cooled...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com