Pre-amplification method for trace DNA applied in medicolegal expertise
A forensic identification and pre-amplification technology, which is applied in the fields of anthropology, forensic evidence biology, and criminal investigation, can solve problems such as difficulties, lost typing, and allelic imbalance, and achieve low-cost effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
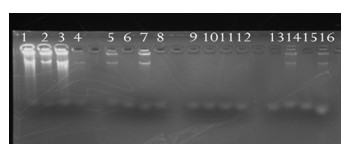
[0050] Embodiment 1: Primer Mix amplifies the method of 100pg, 50pg and 10pg DNA template under 4 gradients, and specific content is as follows:
[0051] 1. The preparation of the primer Mix mixture, the primer Mix mixture is a mixture of 36 primers from primer1-primer36, prepare 4 gradient primer Mix mixtures, the concentration gradient of the mixture is as follows:
[0052] Primer Mix 1: primer1 (final concentration 3uM) and primer2 (final concentration 3uM), primer3-36 (each primer final concentration 0.1uM)
[0053] Primer Mix 2: primer1 (final concentration 3uM) and primer2 (final concentration 3uM), primer3-36 (each primer final concentration 0.05uM)
[0054] Primer Mix 3: primer1 (final concentration 3uM) and primer2 (final concentration 3uM), primer3-36 (each primer final concentration 0.01uM)
[0055] Primer Mix 4: primer1 (final concentration 3uM) and primer2 (final concentration 3uM), primer3-36 (each primer final concentration 0.005uM)
[0056] 2. Add the followi...
Embodiment 2
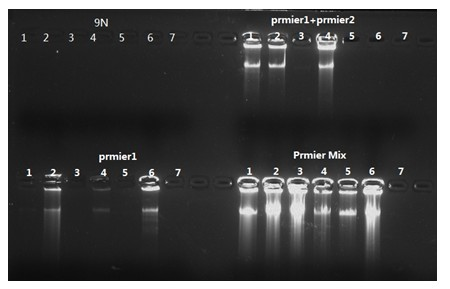
[0066] Embodiment 2: the method for amplifying 100pg, 50pg and 10pg DNA using random primer 9N, primer1 and primer2 double primers, primer1 single primer and Pmier Mix, the specific contents are as follows:
[0067] 1. Random primer 9N amplifies 100pg, 50pg and 10pg DNA, the amplification system is as follows:
[0068] 9N 3ul
[0069] 10×Reaction Buffer 2ul
[0070] 10 (or 50 or 100) pg / ul DNA 1ul
[0071] wxya 2 O 7ul
[0072] Pre-denature at 98°C for 10 minutes, take out the deformed solution, place it on ice quickly for 15 minutes, then add the following reagents to it, and finally amplify at 33°C for 16 hours, 65°C for 15 minutes; 4°C∞;
[0073] Each 2.5mM dNTP 4ul
[0074] 0.5% (w / v) BSA 2ul
[0075] 5000U Phi29 1ul
[0076] figure 2 In "9N", the initial template amount of 1-2 is 100pg, 3-4 is 50pg, 5-6 is 10pg, and 7 is a negative control; no amplification band is shown in the figure.
[0077] 2. Primer 1 (final concentration 3uM) single primer amplifies 100pg,...
Embodiment 3
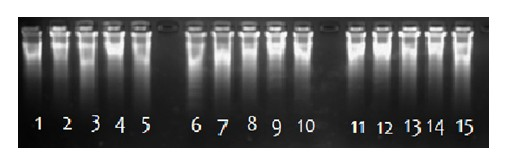
[0109] Embodiment 3: Applied to the forensic identification trace DNA pre-amplification method, using Pmier Mix to amplify the method of 100pg, 50pg and 10pg DNA, the specific content is as follows:
[0110] (1) Put 36 kinds of primers from primer1-primer36 into a single PCR tube and mix evenly. The final concentration of primer1 and primer2 is 1uM, and the final concentration of each primer from primer3-primer36 is 0.05μM. Divide into three groups;
[0111] (2) Add 10 μL of 10× reaction buffer, 42 μL of double distilled water and 1 μL of 10, 50 or 100 pg / ul trace DNA template to each group of mixed solutions;
[0112] (3) Place the mixture in step (2) in a PCR instrument for pre-denaturation at 98°C for 5 minutes;
[0113] (4) Take out the denatured solution, place it on ice for 10 minutes, then add 6 μl of dNTP, 3 μl of 0.5% BSA, and 2uL of 5000U Phi29 DNA polymerase, mix well and put it back into the PCR instrument for amplification. 2.5mM, PCR amplification conditions are...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com