Primer, kit and method for determining lung cancer gene mutation site based on high-flux sequencing technology
A high-throughput, kit-based technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/testing, etc., can solve the problem of difficult gene mutation sites in lung cancer, many false positives and false negatives, poor repeatability, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
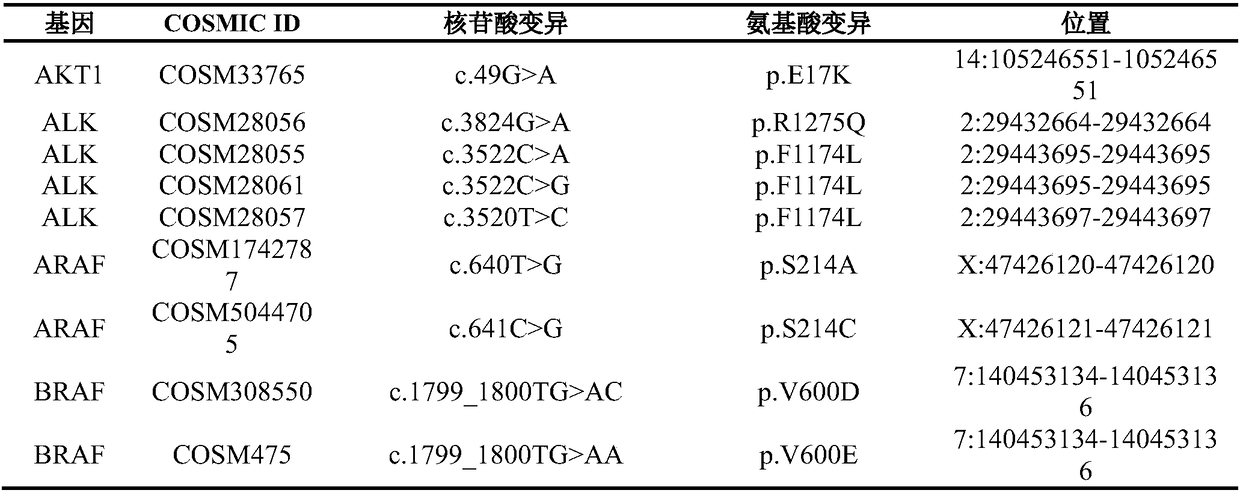
[0063] To detect the biopsy tissue of a certain T2bN1M1 lung squamous cell carcinoma to determine the relevant information of the gene mutation, the steps are as follows:
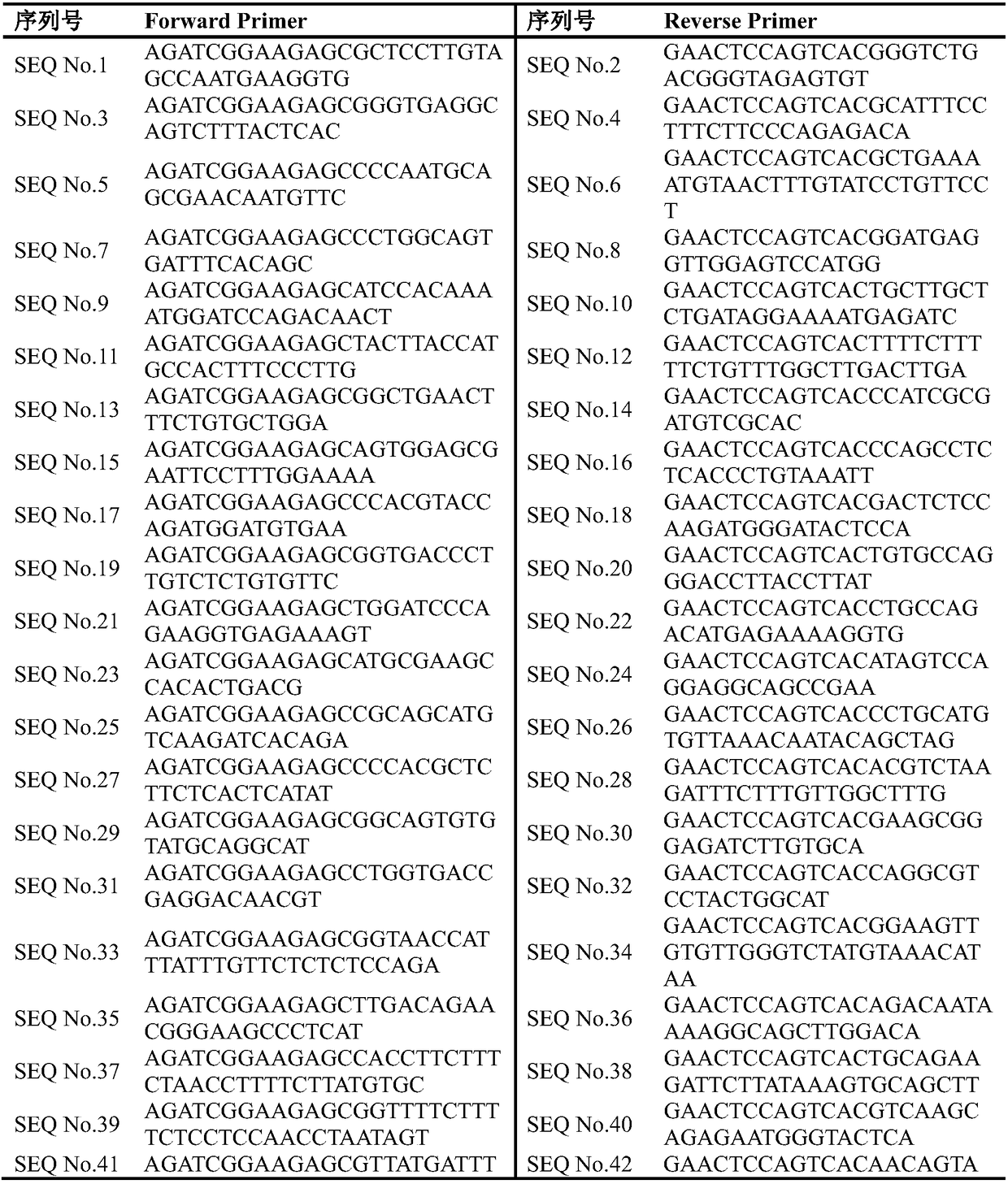
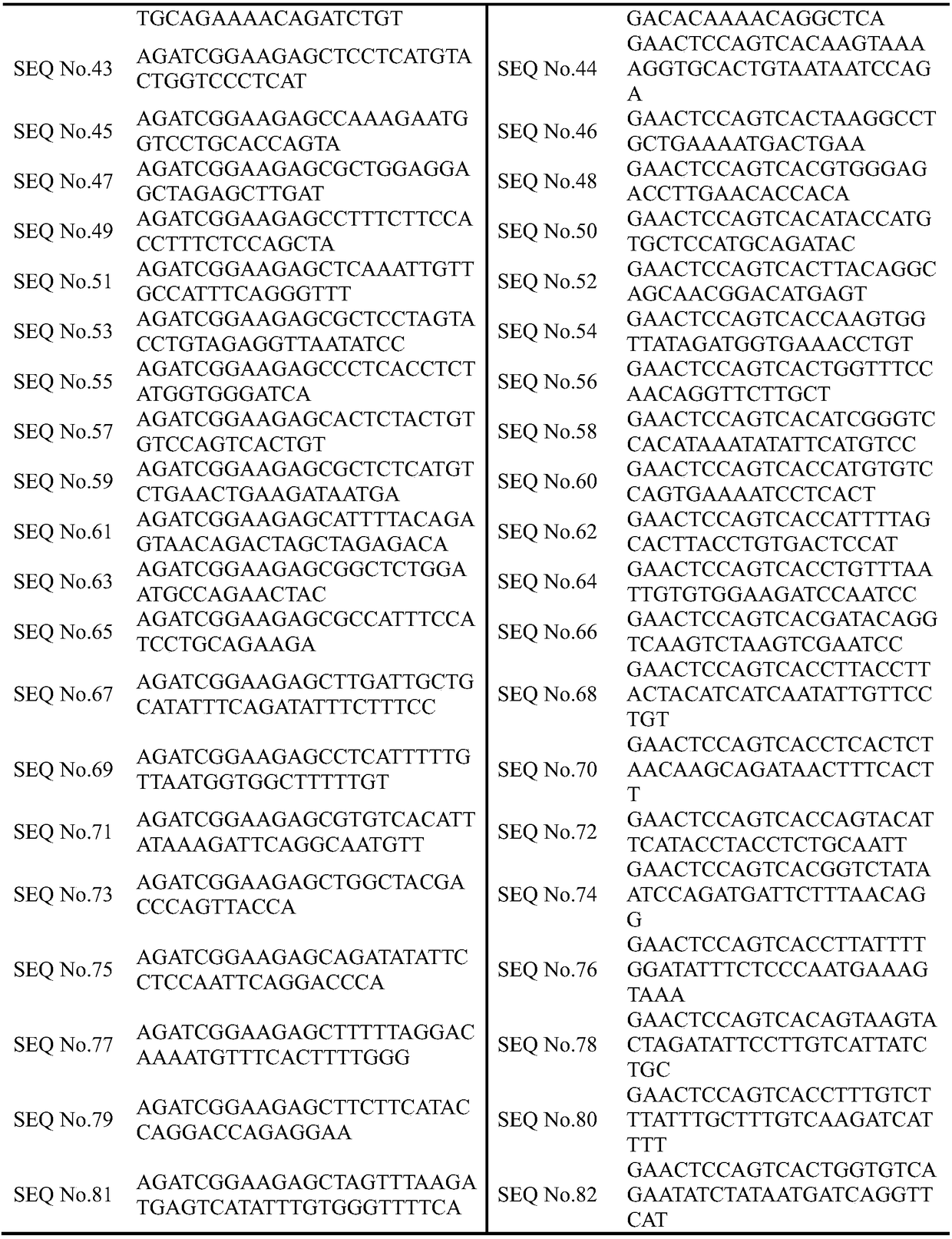
[0064] The sample used was fresh biopsy tissue of T2bN1M1 lung cancer, and the genomic DNA in the fresh biopsy tissue of lung cancer was extracted using a DNA extraction kit, and the extracted genomic DNA was amplified by multiplex PCR using SEQ No. The NextSeq 500 sequencer performs sequencing analysis, and the specific implementation method is as follows:
[0065] A. Genomic DNA extraction: Use the Qiagen DNeasy Blood&Tissue Kit to extract genomic DNA according to the kit instructions. The obtained genomic DNA is subjected to gel electrophoresis and quality detection using NanoDrop 2000, which requires that the genomic DNA has no obvious degradation, the concentration is greater than 10ng / μL, A260 / A280>1.8, A260 / A230>2.0, and Qubit 3.0 is used for accurate quantification.
[0066] B. Multiplex PCR amplif...
Embodiment 2
[0079] A metastatic lung adenocarcinoma was detected by FISH as EGFR copy number amplification and ERBB2 copy number amplification. Using the method of the present invention to carry out molecular detection to determine the specific mutation site.
[0080] The sample used was paraffin-embedded lung cancer tissue, and the genomic DNA in the paraffin-embedded tissue was extracted using a DNA extraction kit, and the extracted genomic DNA was amplified by multiplex PCR using SEQ No.1 to SEQ No.82, and then analyzed by Illumina The NextSeq 500 sequencer performs sequencing analysis, and the specific implementation method is as follows:
[0081] A. Genomic DNA extraction: Genomic DNA was extracted using Qiagen GeneRead DNA FFPE Kit and referring to the kit instructions. The quality of the obtained genomic DNA was detected by NanoDrop 2000, and the DNA concentration was required to be greater than 5ng / μL, A260 / A280>1.8, A260 / A230>2.0, and Qubit 3.0 was used for accurate quantificati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com