Molecular detection method for rapidly identifying carbendazim-resistant genotype-F200Y botrytis cinerea strain
A technology for Botrytis cinerea and molecular detection, which is applied in the detection field of molecular biology to achieve the effects of high accuracy, strong specificity and improved detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1 LAMP reaction system optimization
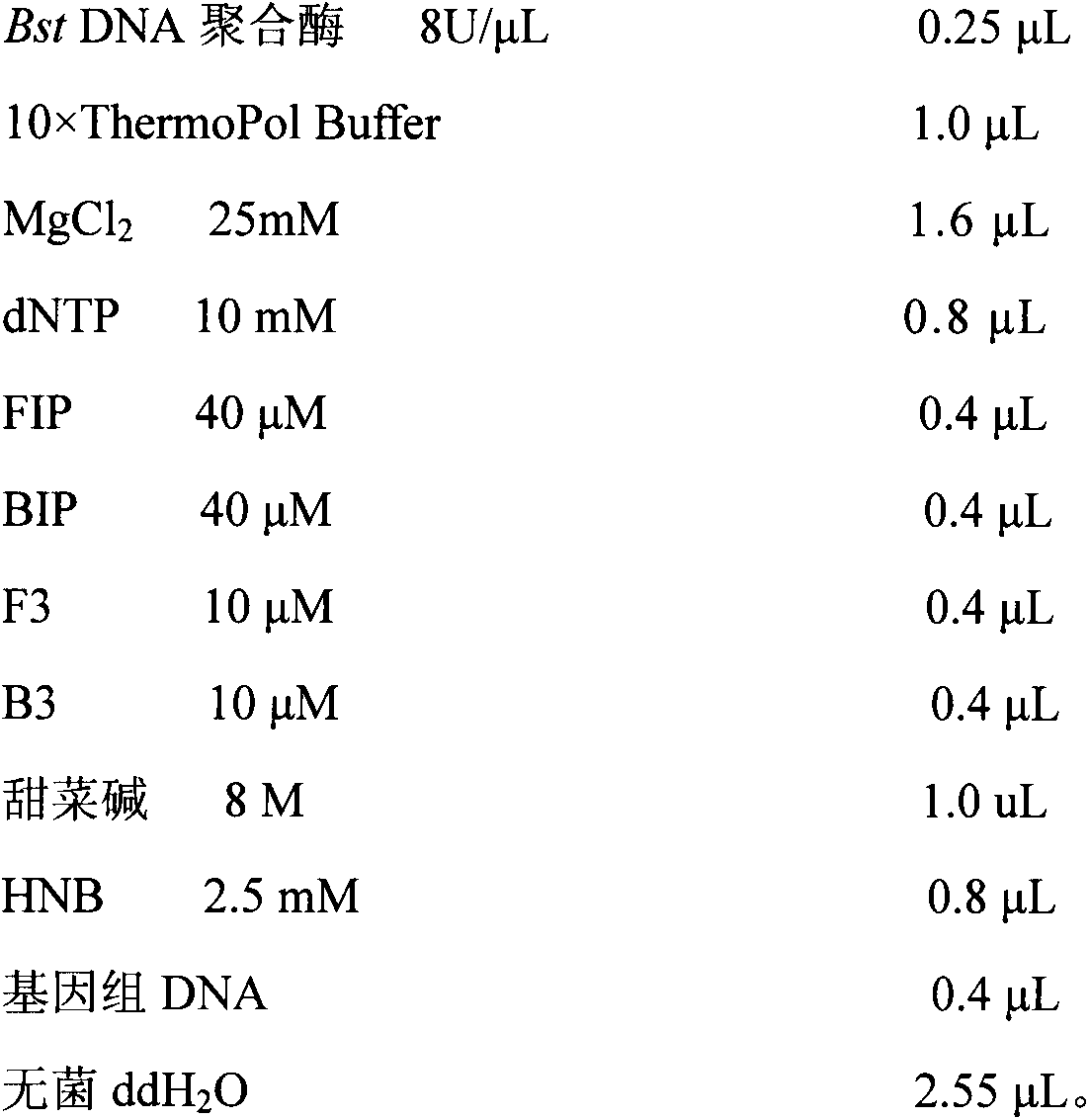
[0031] In order to save the identification cost and ensure the stability and reliability of the identification method, BstDNA polymerase (8U / μL) (0.8-4.0U), Mg 2+ (25mM) concentration (0.6-2.0μL), primer FIP / BIP (40μM) and F3 / B3 (10μM) concentration (0.1-0.5μL), betaine (8M) concentration (0.4-2.0μL), HNB (2.5mM ) concentration (0.4-1.2μL) was optimized, and the best reaction system was determined to be: BstDNA polymerase (8U / μL) 0.25μL, 10×ThermoPol 1μL, MgCl 2 (25mM) 1.6μL, dNTP (10mM) 0.8μL, FIP (40μM) 0.4μL, BIP (40μM) 0.4μL, F3 (10μM) 0.4μL, B3 (10μM) 0.4μL, betaine (8M) 1.0μL, HNB (2.5mM) 0.8μL, Genomic DNA 0.4μL, sterile ddH 2 O 2.55 μL.
Embodiment 2
[0032] Example 2 LAMP reaction parameter optimization
[0033] In order to obtain the optimum reaction temperature and time and ensure the high efficiency of the detection method, the reaction temperature and time in the reaction parameters were optimized, and the optimum reaction temperature and time were obtained to be 60°C and 45min, respectively.
Embodiment 3
[0034] Embodiment 3 LAMP reaction sensitivity test
[0035] In order to determine the lower limit of detection of the LAMP reaction, clone the DNA fragment containing the 200-position mutation site of Botrytis cinerea to the carbendazim-resistant genotype F200Y strain into the vector pEASY-T3, transform Escherichia coli, and pick positive transformants, After the plasmid was extracted, the 10-fold serial dilution was used as a template for LAMP and PCR amplification respectively. Finally, it was concluded that the lowest detection limit of LAMP was 100 times that of ordinary PCR.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com