Method for measuring short chain RNA by amplifying length polymorphism of DNA fragment
Through the amplified DNA fragment length polymorphism quantitative determination method, the use of internal standards and dynamic small RNA standards without natural homologous sequences solves the sensitivity and standardization issues of miRNA quantitative determination and achieves highly accurate and comparable results. Suitable for clinical diagnosis and research of complex biological samples.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
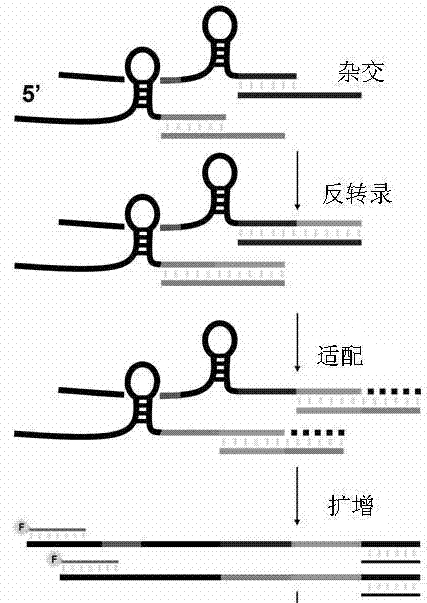
[0060] Embodiment 1 miRFLP quantitative analysis method reaction process and measurement principle
[0061] This example takes omega primers as an example to specifically illustrate the reaction process and measurement principle of the miRFLP quantitative analysis method.
[0062] The miRFLP reaction consists of four steps: miRNA reverse transcription, cDNA tailing, PCR synchronous amplification, and fluorescent fragment length polymorphism analysis of PCR products, such as Figure 1A and Figure 1B shown. The first step of the reaction is to hybridize miRNA and omega primers, miRNA is paired with complementary probes, and then reverse transcriptase is used to synthesize cDNA using the unpaired miRNA 3' end as a template. After removing the RNA, the newly synthesized cDNA is hybridized with a 3' oligonucleotide primer containing a common PCR target and DNA polymerase is used to fill the single-stranded gap after DNA pairing starting from the 3' end of the cDNA. After this co...
Embodiment 2
[0064] Example 2 Determination of Fluorescent Intensity of PCR Fluorescently Labeled Product Equal Dilutions
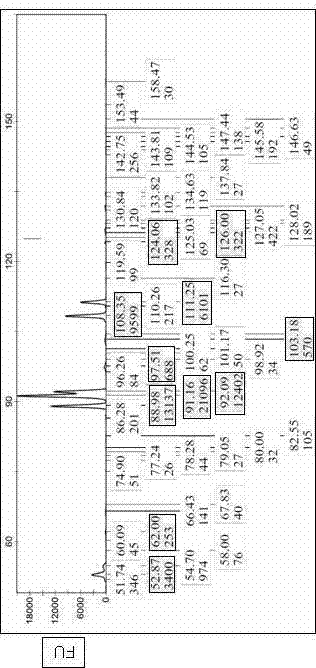
[0065] The fluorescence intensity measured by the fluorescence quantitative analyzer is directly proportional to the amount of the fluorescent substance to be measured. This relationship is related to the fluorescence probe configured by the instrument, that is, different fluorescence probes have different fluorescence response curves. The fluorescence intensity measured by ABI's Prizma 310 DNA sequencer has a linear and quantitative relationship with the amount of fluorescent substance to be tested in the range of 5-7000 FU, and changes to a parabola when the fluorescence intensity value is greater than 7000FU. The response characteristics of fluorescence probes of different instruments are different, which can affect the regression relationship between the measured fluorescence intensity and the measured fluorescence amount. In this embodiment, the fluorescence resp...
Embodiment 3
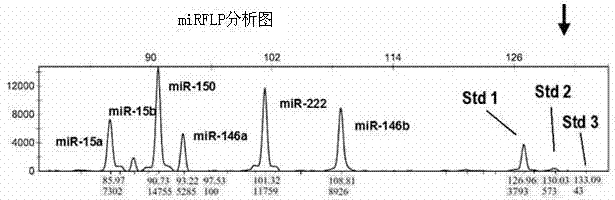
[0067] Example 3 miRFLP quantitative analysis assay test of miR-92a and miR-92b
[0068] In the miRFLP analysis method, the miRNA to be tested and the dynamic miRNA standard are evenly mixed, and after miRNA reverse transcription, cDNA tail modification and fluorescent PCR synchronous amplification, DNA sequencer is used to analyze the length of DNA fragments and quantitative fluorescence. Specifically, first prepare 4 μl hybridization master mix: 2 μl 5xRT buffer, 1 μl 10 mM MgSO 4 , 1 μl dynamic miRNA standard mixture, the mixture includes the number of molecules is 3 × 10 6 The "Standard 1 RNA" (i.e. Figure 4A "std1", the same below), the number of molecules is ×10 5 The "Standard 2 RNA" (i.e. Figure 4A "std2", the same below), the number of molecules is 3×10 4 The "Standard 3 RNA" (i.e. Figure 4A "std3" in , the same below), the RNA sequence of the dynamic miRNA standard is shown in Table 2, and the sequence search of the Sangers miRbase20 database proves that ther...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com