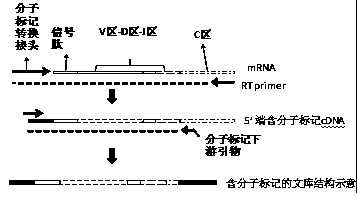
Construction method of immune repertoire sequencing library and test kit
A technology for sequencing libraries and immunological libraries, applied in chemical libraries, biochemical equipment and methods, combinatorial chemistry, etc., can solve the problems of inability to completely eliminate PCR amplification bias, prone to amplification bias, poor specificity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 Application of a Human TCR Diversity Detection Kit on the Illumina Sequencing Platform
[0058] 1. Extraction of total RNA.
[0059] 1. Add 5 times the volume of 1X red blood cell lysate H to 200uL of human whole blood.
[0060] 2. Incubate on ice for 10-15 min, and vortex to mix twice during the incubation.
[0061] 3. Centrifuge at 2,100 rpm (~400×g) for 10 min at 4°C to remove the supernatant completely.
[0062] 4. Add 1X Red Blood Cell Lysis Buffer H to the white blood cell pellet to resuspend the cells.
[0063] 5. Centrifuge at 2,100 rpm (~400×g) for 10 min at 4°C to remove the supernatant completely.
[0064] 6. Add 350uL Lysis Solution RLH (please add β-mercaptoethanol before use) to the white blood cell pellet, vortex or use a pipette to mix.
[0065] 7. Transfer the solution to the filter column CS (the filter column CS is placed in a collection tube), centrifuge at 12,000 rpm (~13,400×g) for 2 min, discard the filter column CS, and collect the ...
Embodiment 2
[0093] Example 2 Application of a Human BCR Diversity Detection Kit on the Illumina Sequencing Platform
[0094] 1. Extraction of total RNA.
[0095] 2. Reverse transcription.
[0096]1. Take 100ng total RNA, add IGHCo primer (SEQ ID No.11), IGKCo primer (SEQ ID No.12), IGLCo primer (SEQ ID No.13), dNTPs, reverse transcription Buffer, reverse transcriptase, ILTS linker (SEQ ID No.5), total system 20uL.
[0097] 2. React on PCR machine, 50°C for 30 minutes; 25°C for 5 minutes; 42°C for 25 minutes.
[0098] 3. Pre-amplification.
[0099] 1. Take 2uL reverse transcription product, add PCR master mix, ILT primer (SEQ ID No.6), IGHCi primer (SEQ ID No.14), IGKCi primer (SEQ ID No.15), IGLCi primer (SEQ ID No. 16), add water to 20uL system.
[0100] 2. React on a PCR machine, 95°C for 3 minutes; 95°C for 30 seconds, 65°C for 30s, 72°C for 30s, 20 cycles; 72°C for 5 minutes.
[0101] 3. Add 30uL kapa magnetic beads and mix well, let stand at room temperature for 5 minutes, sepa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com