Potato low-temperature sweetening resistant molecular marker combination and application thereof in potato low-temperature sweetening resistant breeding
A technology of molecular markers and low-temperature saccharification, which is applied in the fields of molecular biology and genetic breeding, can solve the problems of difficult to achieve ideal results and increase the difficulty of breeding, and achieve the effects of easy detection, high sensitivity and good accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
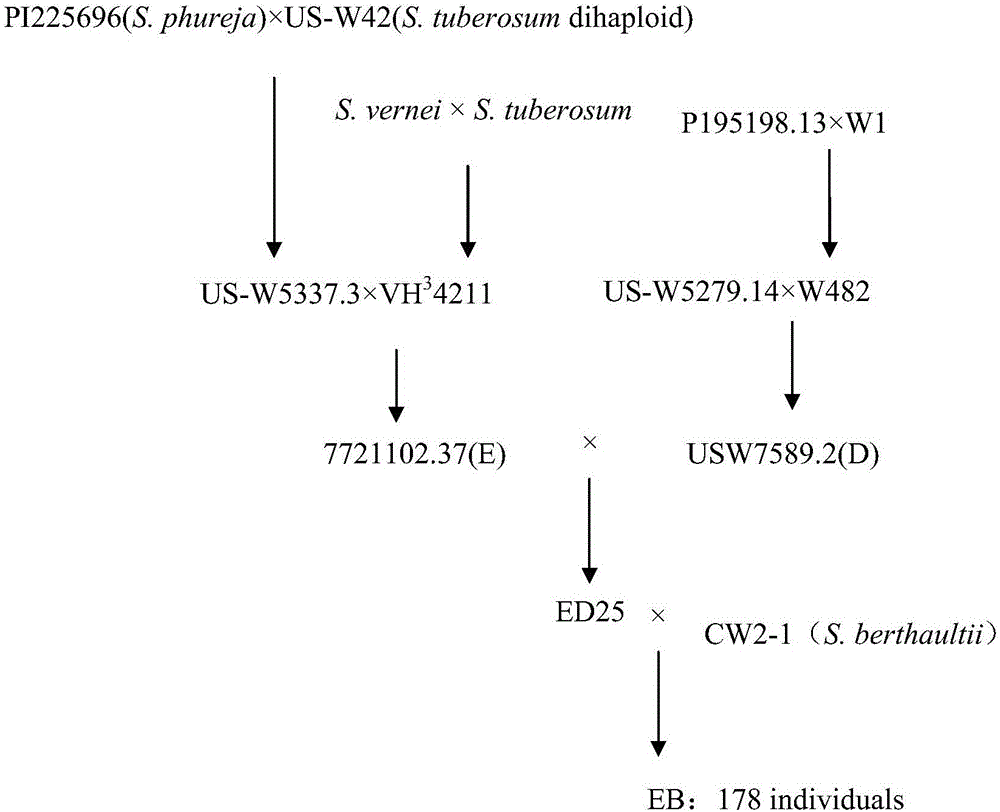
[0037] 1. Materials: The diploid potato F1 population (EB population) is used as the material. The EB population is 178 offspring produced by crossing the female parent ED25 and the male parent CW2-1. For details, see figure 1 shown. Among them, ED25 contains the blood of two cultivated species S.phureja, S.tuberosum and one wild species S.vernei, which is not resistant to low-temperature saccharification; CW2-1 is a clone of the wild potato species S.berthaultii, which is resistant to low-temperature saccharification.
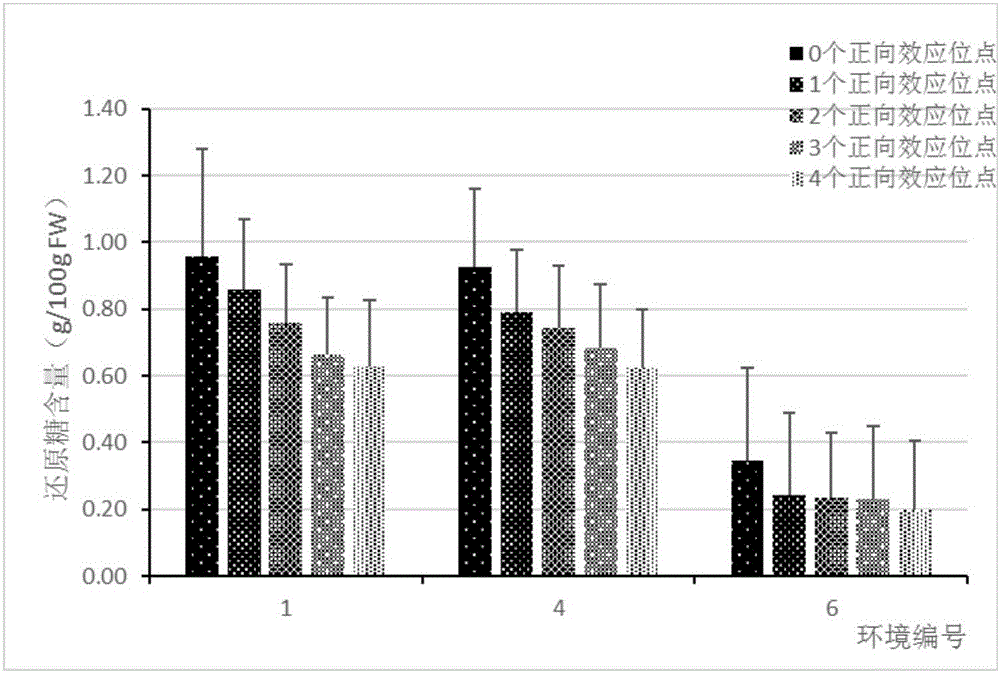
[0038] The results of phenotypic identification of EB population showed that the reducing sugar content of offspring after low-temperature storage and after warming was significantly separated, showing a normal distribution, which was suitable for QTL mapping analysis.
[0039] 2. Molecular marker experiment method
[0040] 2.1 AFLP markers
[0041] 1) Extraction of genomic DNA
[0042] Genomes were extracted from the young leaves of the EB population using...
Embodiment 2
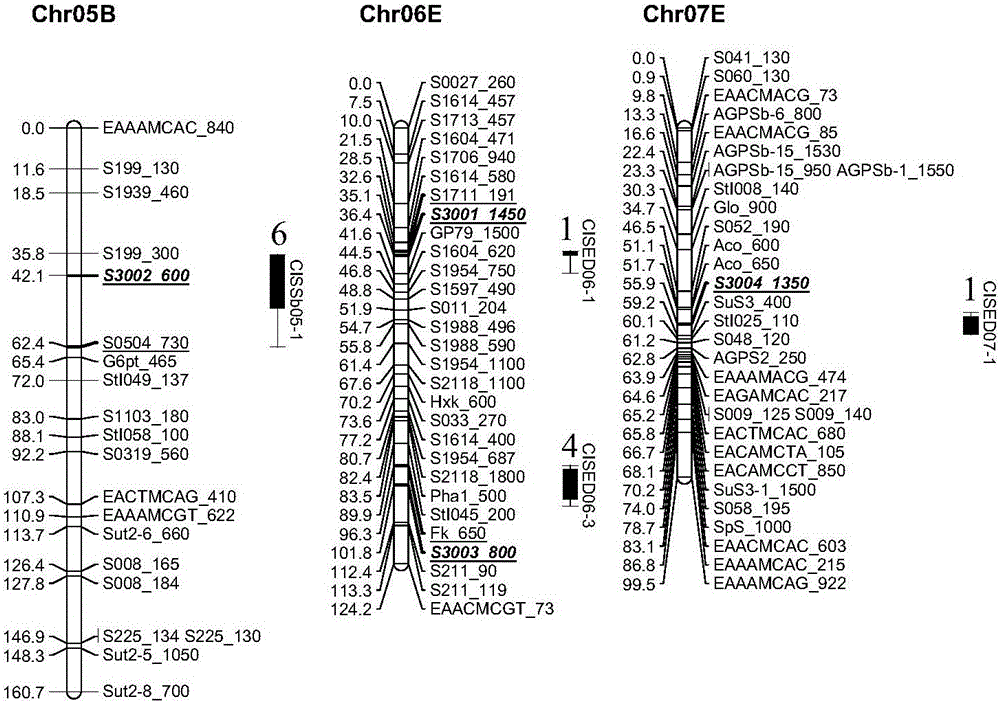
[0096] Verify the 4 molecular markers screened in Example 1
[0097] The present invention measures the EB population after being planted in 3 different environments (Changling Hill (109.8E, 30.2N), Tianchi Mountain (109.7E, 30.3N) and Wuhan (114.4E, 30.5N)) After the tubers were stored at low temperature ( 4° C. 30 d), the reducing sugar content determination method is the same as that in Example 1; the field experiment design adopts the sequential arrangement method, and each genotype has 3 repetitions, and 10 plants are planted in each repetition.
[0098] Genomes were extracted from young leaves of the EB population using the improved CTAB method, and the specific steps were the same as in Example 1.
[0099] After detection, the content of the extracted genome is 50-200ng / μl, and then the primer pair of S3001-S3004 nucleic acid sequence is used for landing PCR.
[0100] The drop-down PCR system of each molecular marker is 20 μl, specifically as follows: ultrapure water (...
Embodiment 3
[0108] The primer pairs of S3001-S3004 obtained in Example 1 were respectively verified for the broadness of low-temperature glycation resistance.
[0109] 64 potato varieties, breeding parents and breeding high-generation lines with different low-temperature saccharification resistance were selected. The above-mentioned 64 natural population materials were planted in the potato greenhouse of the Huazhong Sub-center of the National Vegetable Improvement Center of Hubei Province. Each material was planted in 4 pots. After the mixed harvest, each material picked 4 larger tubers without pests and diseases and kept them at low temperature ( 4° C.) After storing for 30 days, the reducing sugar content in each tuber was measured by the method in Example 1.
[0110] Genomes were extracted from young potato leaves planted with different materials according to the method in Example 1. The content of the extracted genomes was 50-200 ng / μl, and then landing PCR was performed with primer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com