Method for evaluating DNA quality of FFPE sample
A sample medium and high-quality technology, applied in the field of molecular biology, can solve the problems of poor quality, small number of FFPE samples, single detection target area, etc., and achieve the effect of accurate DNA quality grading, clear result bands, and multiple detection positions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Evaluation experiment of DNA fragment quality in FFPE samples
[0050] 1. Primer Preparation
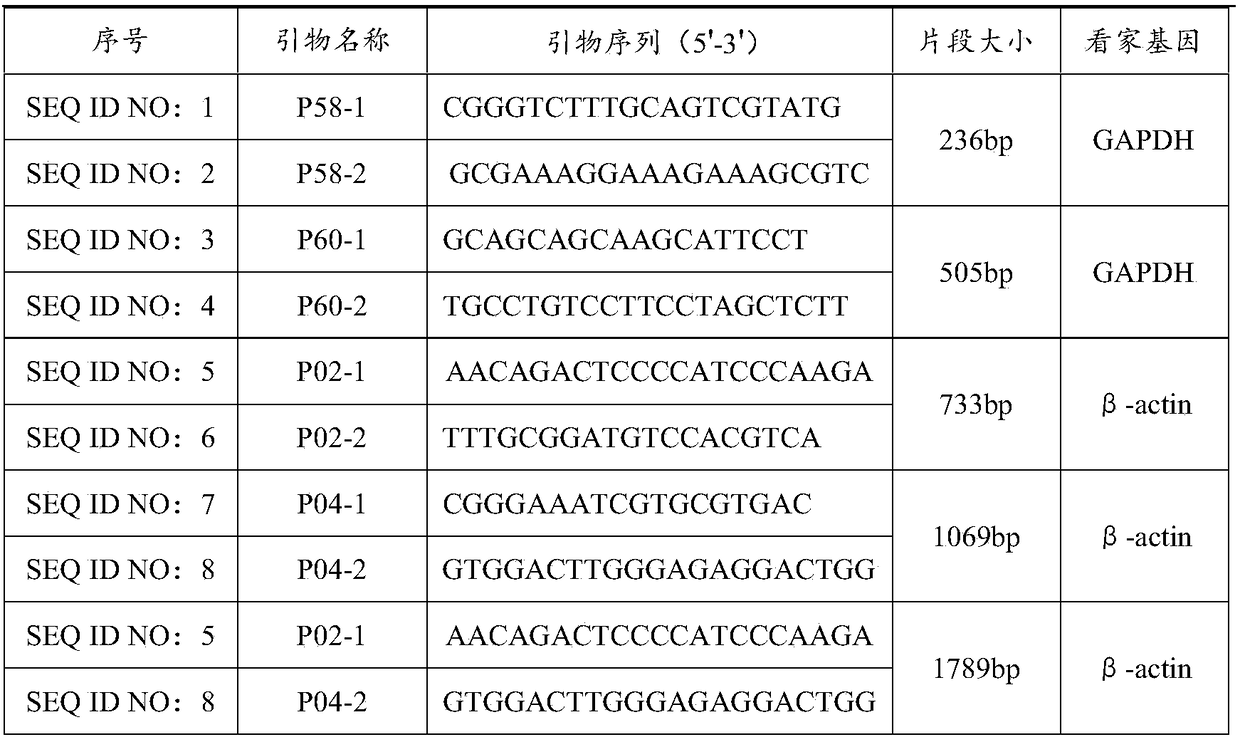
[0051] According to the sequence of the GAPDH gene as a template, primers P58-1, P58-2, P60-1 and P60-2 were designed, wherein, primers P58-1 and P58-2 were designed to amplify a fragment with a length of 236bp, and primer P60 -1 and PP60-2 were used to amplify a fragment with a length of 505bp. According to the sequence of the β-actin gene as a template, primers P02-1, P02-2, P04-1 and P04-2 were designed, wherein, the obligatory P02-1 and P02-2 were designed to amplify a fragment with a length of 733bp, Primers P04-1 and P04-2 were designed to amplify a fragment with a length of 1069bp, and primers P02-1 and P04-2 were designed to amplify a fragment with a length of 1789bp. The nucleotide sequences of the above primers are shown in Table 1.
[0052] Table 1
[0053]
[0054] 2. DNA extraction from FFPE samples
[0055] FFPE samples with storage times of 0.5...
Embodiment 2
[0071] Sample 1, sample 2, and sample 3 of Example 1 were used for database construction.
[0072] (1) Preparation of DNA fragments to be sequenced
[0073] Take sample 1, sample 2, and sample 3 500ng each for DNA fragmentation. The fragmentation condition is 30 cycles, 30 seconds ON, 30 seconds OFF, fragmented to a fragment size of about 200bp, and the double-stranded DNA fragment after fragmentation Some of these are blunt-ended, others contain 3' or 5' overhangs.
[0074] (2) Perform end repair to obtain blunt-ended DNA fragments.
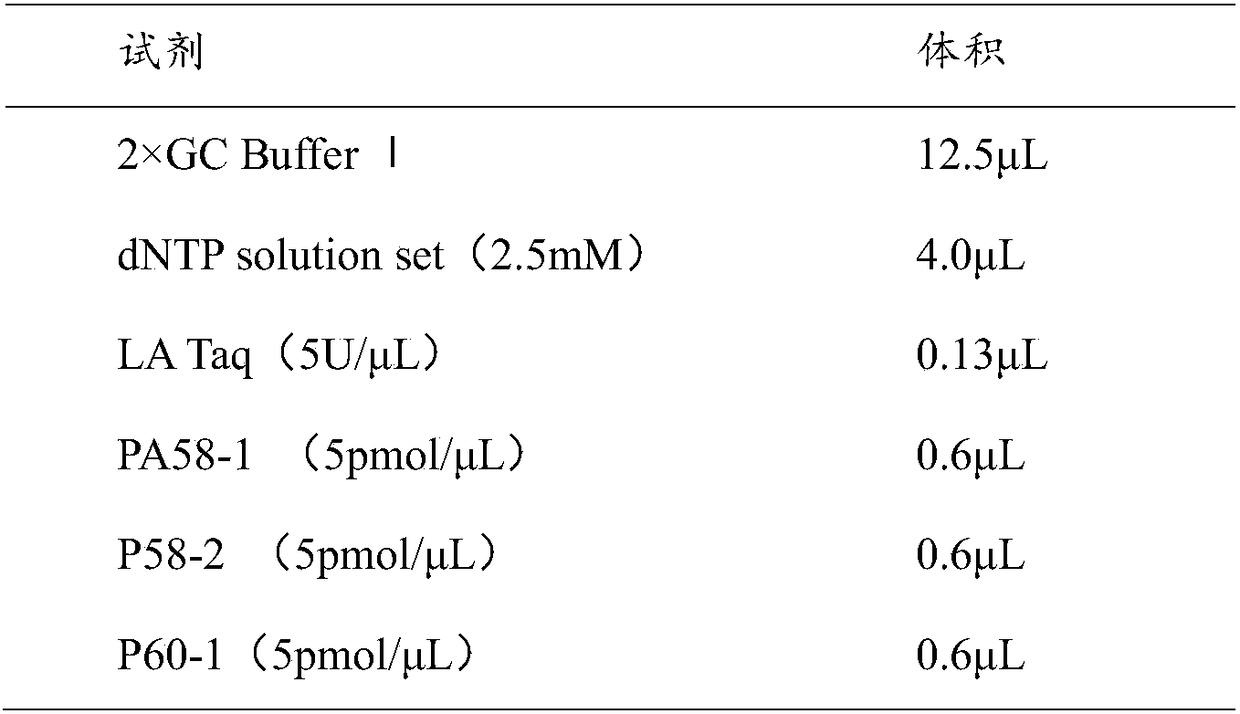
[0075] A. The reaction system is as follows:
[0076]
[0077]
[0078] B. Incubate in a PCR instrument for 30 minutes at a temperature of 20°C;
[0079] C. The reaction product was purified with magnetic beads, and eluted with 19.5 μL of elution buffer EB (Elution Buffer) to obtain blunt-ended DNA fragments.
[0080] (3) Add A to the blunt-ended DNA fragments at the 3' end
[0081] A. The reaction system is as follows:
[0082]
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com