Identification primer suitable for hippopus and common tridacna varieties in South China Sea and hybrids thereof and method thereof
A technology for hybrids and Tridacna, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as difficulty in identification of morphological characteristics and identification in the early stage of development, and achieve intuitive and accurate results. Simple method and low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
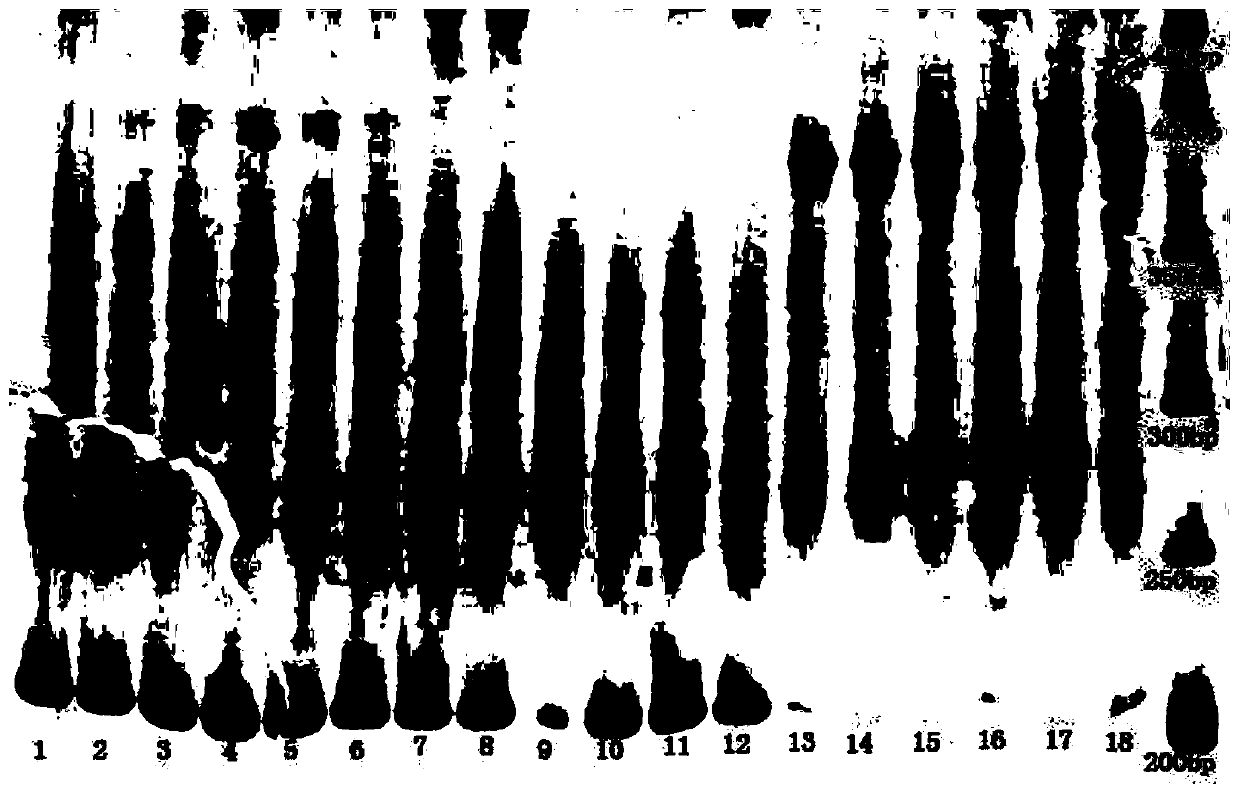
[0017] see figure 1 , using phenol-chloroform extraction method to extract the genomic DNA of 40 individuals of giant clam oyster, scaleless giant clam, scaled giant clam, long giant clam, safflower giant clam and nova giant clam. The specific method is to cut the adductor muscle fully, absorb the water with clean filter paper, put it into a 1.5mL centrifuge tube, then add 400μl lysate and 10μl proteinase K (10mg / ml), mix well on the shaker, 55 Digest in a water bath at ℃ for 3-5 hours until the lysate is clear. Add an equal volume of saturated phenol (200 μL), chloroform / isoamyl alcohol (24:1) (200 μL) mixture and extract three times. Precipitate DNA with 1 mL of absolute ethanol, wash with 70% alcohol, dry at room temperature and dissolve in 100 μL of ultrapure water. The concentration and purity of DNA were detected by ultraviolet spectrophotometer, and the integrity of DNA was detected by 1% agarose gel electrophoresis. The genome DNAs of oyster, scaleless, scaled, long...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com