Microsatellite primers for identifying crassostrea hongkongensis, crassostrea ariakensis and crassostrea gigas and hybrid thereof and identification method
A Pacific oyster and microsatellite technology, applied in the field of microsatellite primers and identification, can solve the problems of high cost, small differences in gene sequences, differentiation, etc., and achieve the effects of low cost, intuitive results, and simple methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
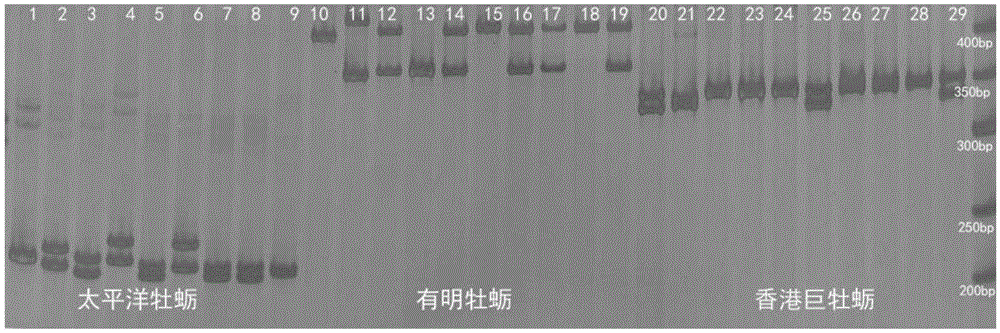
[0019] Genomic DNA of 90 individuals of Hong Kong giant oyster, Ariake oyster, and Pacific oyster were extracted by phenol-chloroform extraction, and the specific methods were as follows:
[0020] Fully cut up the adductor muscle, absorb the water with clean filter paper, put it into a 1.5mL centrifuge tube, then add 400μl lysate and 10μl proteinase K (10mg / ml), mix well on a shaker, and digest in a water bath at 55°C 3 to 5 hours until the lysate is clear. Add an equal volume of saturated phenol (200 μL), chloroform / isoamyl alcohol (24:1) (200 μL) mixture and extract three times. Precipitate DNA with 1 mL of absolute ethanol, wash with 70% alcohol, dry at room temperature and dissolve in 100 μL of ultrapure water. The concentration and purity of DNA were detected by ultraviolet spectrophotometer, and the integrity of DNA was detected by 1% agarose gel electrophoresis. The genomic DNAs of Hong Kong giant oyster, Ariake oyster, and Pacific oyster were obtained respectively. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com