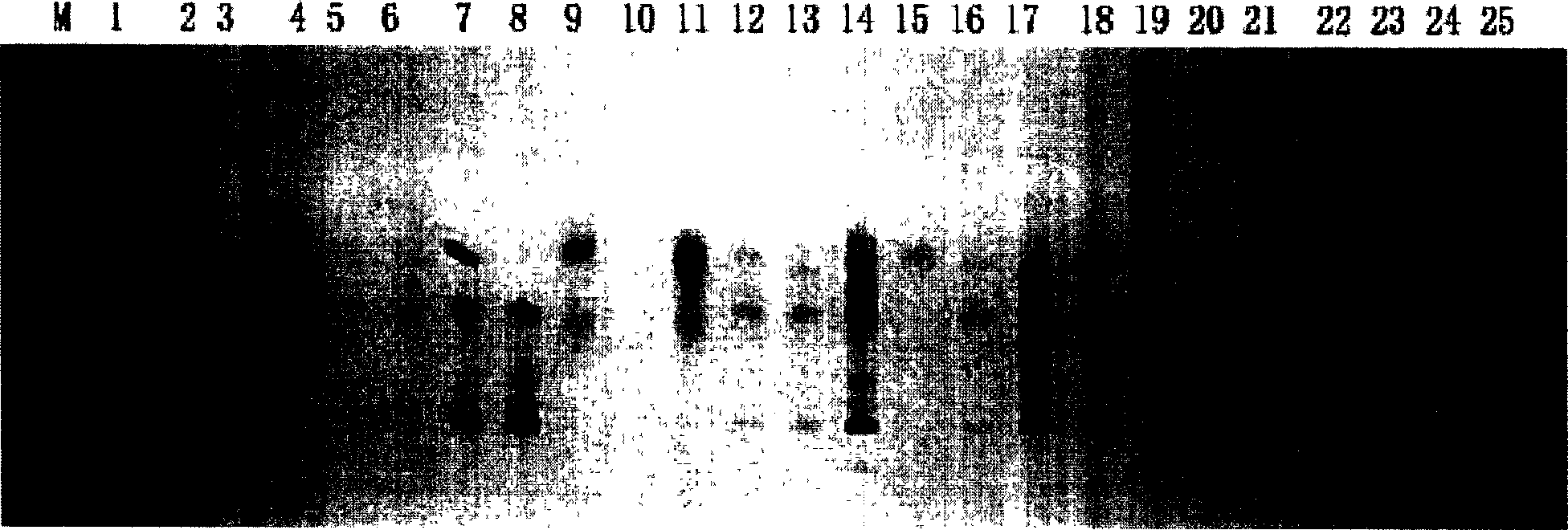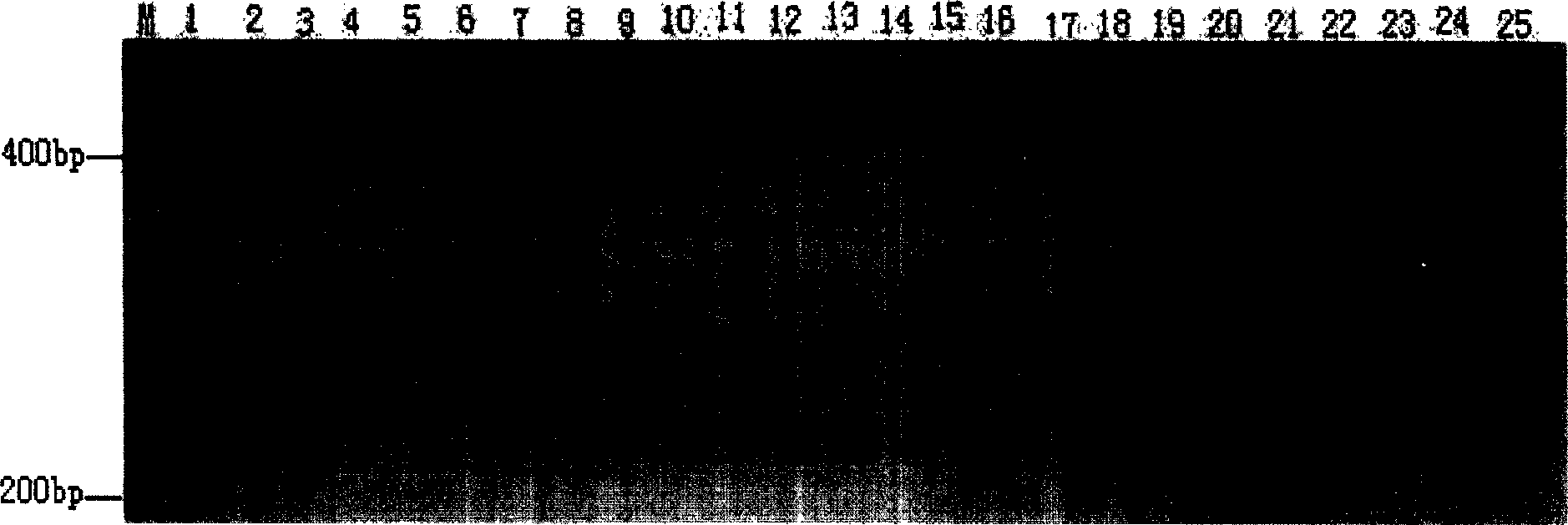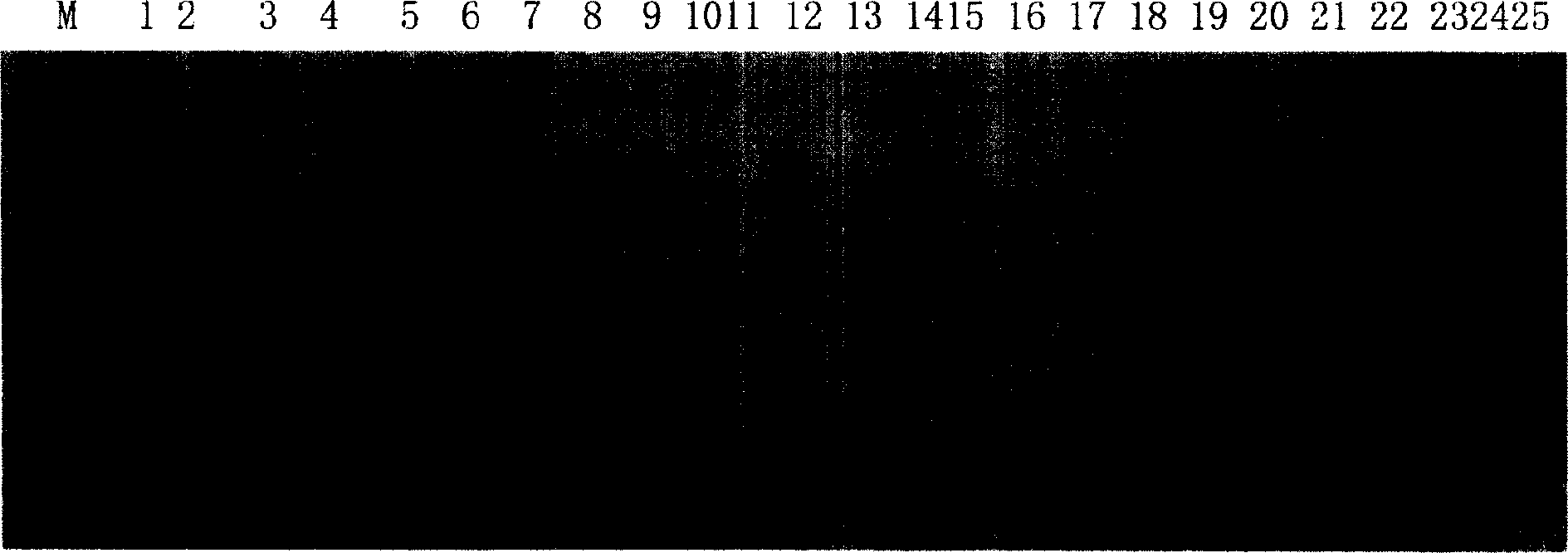Ramie micro-satellite DNA Label
A microsatellite marker, ramie technology, applied in the field of ramie DNA molecular genetic markers, can solve the problems of limited developmental period, low accuracy, and inability to distinguish homozygous genotypes from heterozygous genotypes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0021] 1: Ramie (CT / AG) n Construction of Microsatellite Enriched Library
[0022] (1) According to Guo Anping's method (see Guo Anping, 1997, China Mazuo, 19: 4-10) to extract ramie genomic DNA. Use restriction endonuclease Mse I digestion ramie genome DNA, digestion product is electrophoresis on 1.2% agarose gel, cuts out the DNA fragment of 300-1000bp size under ultraviolet light and uses gel recovery kit (purchased from Qiagen company) Purify the recovered digested product.
[0023] (2) base-complementary oligonucleotide PHC-1 (5'-AGATGGAATTCGTACACTCGT-3') and phosphorylated oligonucleotide PHC-2 (5'-phos-TAACGAGTGTACGAATTCCATCT-3') were mixed equimolarly, Denature at 95°C for 5 minutes and anneal at 60°C for 1 hour to form a linker with MseI sticky ends.
[0024] (3) Ligate the digested product and adapter with T4 DNA ligase. The ligated product was extracted once with an equal volume of phenol / chloroform / isoamyl alcohol (25:24:1), and the supernatant was collected by...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com