Method for detecting Jassr131 microsatellite deoxyribonucleic acid (DNA) marker of Charybdis japonica
A DNA labeling and detection method technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems that have not yet been seen, and achieve the effect of simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
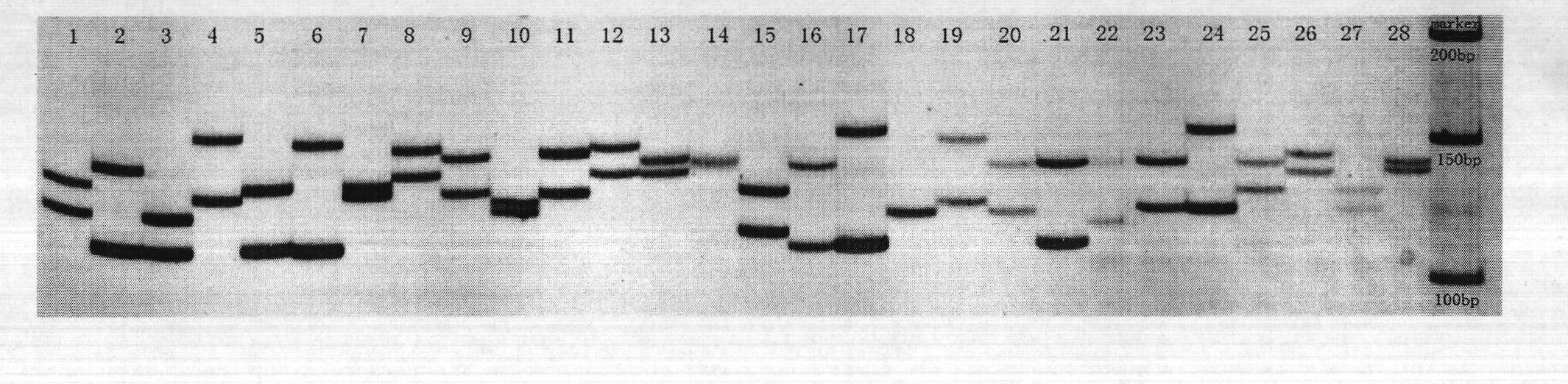
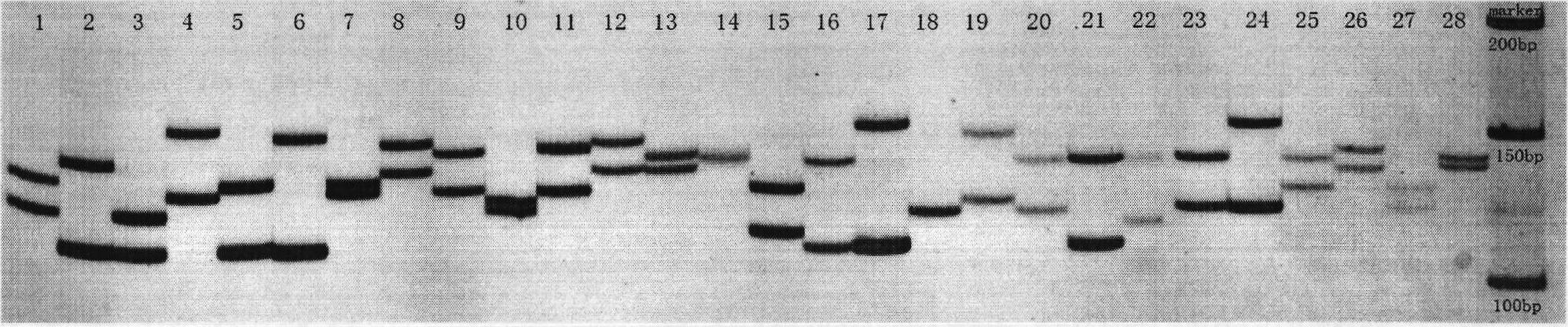
[0016] The following examples describe in detail the present invention's DNA molecular genetic marker technology of Jassr131 microsatellite core sequence. Firstly, extract the genomic DNA of Japanese catfish and dilute it for later use; then use the Jassr131 microsatellite core sequence in the Japanese catfish genome library, and design specific primers at both ends of the sequence; The genomic DNA was amplified by PCR, and the PCR products were detected by polyacrylamide gel electrophoresis; the bands that appeared in the products were analyzed to determine the genotype of each individual, and obtained the polymorphism of the highly genetic variation in the core sequence region of Jassr131 in Jassr131. Morphological map (as shown in Figure 1).
[0017] The core sequence of Jassr131 in Japanese catfish is as follows:
[0018] GATCTT CCAGGGAATTGAAACACT AATTTGGCATTTTAGAGTAAGCTATCTAATTTTTTTTCAGCAAGTCTCCTGTTATTTTATTGATTGATAGATTAGTGCTTGAGTGAGTGAGTGAGTG G G TGAACGAGTGGGTGAGTCAGTG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com