Hibiscus cannabinus L. expression sequence tag SSR (Simple Sequence Repeat) DNA markers
A technique for expressing sequence tags and DNA markers, which can be applied in recombinant DNA technology, DNA/RNA fragments, microbial determination/inspection, etc., and can solve problems such as late start of research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Based on the transcriptome data of kenaf, 24 different types of transcription factor expression sequence tags were determined, and 24 new microsatellite markers of kenaf were developed, as shown in Tables 1 and 2.
Embodiment 2
[0029] Embodiment 2: Utilize the kenaf germplasm resources of different sources, through PCR amplification and electrophoresis detection, determine 24 microsatellite markers with abundant polymorphism, as table 3 and figure 1 shown.
[0030] Table 3 Polymorphism and amplification efficiency of kenaf EST-SSR
[0031]
[0032] ABCD indicates the level of clarity of the electrophoretic band pattern.
Embodiment 3
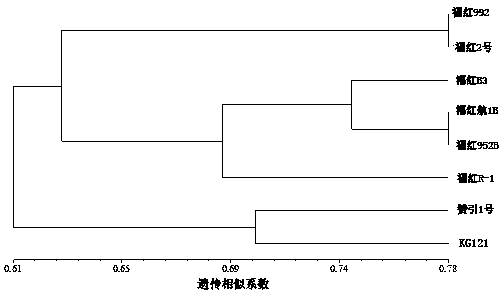
[0033] Example 3: Analysis of genetic diversity of kenaf germplasm resources using the microsatellite markers developed above. The test materials come from the Research Office of Hemp Genetics and Breeding of Fujian Agriculture and Forestry University, namely Fuhong 992, Fuhong 2, Fuhonghang 1B, Fuhong B3, Fuhong R-1, KG121, Zanyin 1 and Fuhong 952B . Among them, Zanyin No. 1 is an excellent strain introduced and bred from Zambia, with red stems, round leaves, and tall plants; while other materials are high-generation strains bred by the research group, with green stems, split leaves, and plants higher. Using NTsys software to calculate the genetic similarity coefficient of 8 kenaf materials. The calculation method is unweighted cluster analysis (UPGMA), and the reading band adopts the method of 0, 1, 0 means no band, and 1 means band. The results of cluster analysis showed that these 24 microsatellite markers could be well used in the genetic diversity analysis of kenaf. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com