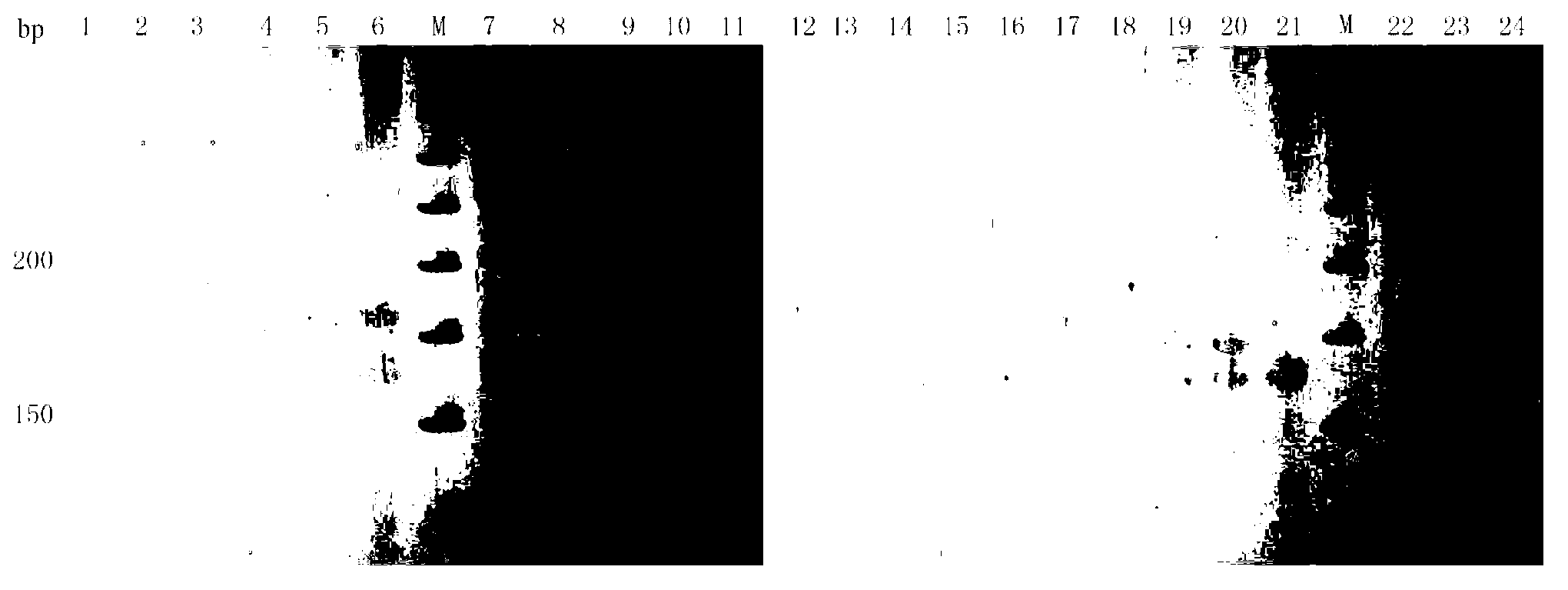Radix polygonati officinalis microsatellite DNA (deoxyribonucleic acid) molecular marker
A DNA molecule, microsatellite marker technology, applied in recombinant DNA technology, DNA/RNA fragment, determination/inspection of microorganisms, etc., can solve the problems of few researches on molecular breeding and low reproduction coefficient of Polygonatum japonica
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
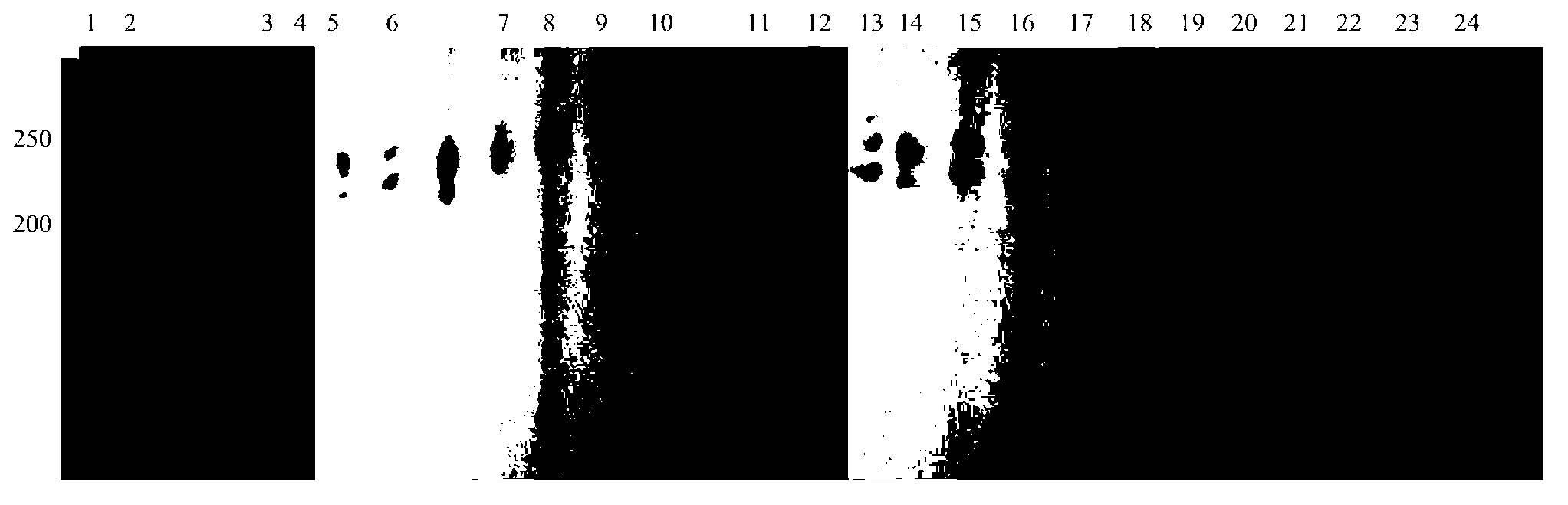
Image
Examples
Embodiment 1
[0024] Construction of Polygonatum Polygonatum Microsatellite DNA Enrichment Library
[0025] 1) Genome extraction and digestion:
[0026] Using the CTAB method introduced by Doyle J (Doyle J. DNA protocols for plants-CTAB total DNA isolation [A]. In Hewitt GM, Johnston A (eds.), Molecular Techniques in Taxonomy [M]. Berlin: Springer, 1991, 283 -293) to extract the genome. The genome was digested with restriction endonuclease Sau 3AI. After 2% agarose gel electrophoresis, the DNA fragments of 300-900 bp were excised under ultraviolet light. Recover with gel recovery kit.
[0027] 2) Add connectors:
[0028] Add the gel recovered and purified product to the adapter:
[0029] oligo A (5'-GGCCAGAGACCCCCAAGCTTCG-3')
[0030] oligo B (5’-PO 4 -GATCCGAAGCTTGGGGTCTCTGGCC-3'),
[0031] Under the action of T4 DNA Ligase, they were ligated overnight in a water bath at 16°C.
[0032] 3) PCR detection adapter connection:
[0033] Use oligo A as primer and ligation product as tem...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com