Method for designing, amplifying and sequencing twelve pairs of floccularia luteovirens microsatellite primers
A microsatellite, curly hair technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA preparation, etc., can solve the problem of reducing heterozygosity, insufficient to complete small-scale spatial genetic structure research, SSR sequence polymorphism It can improve the accuracy, eliminate the interference of invalid alleles, and eliminate the tedious steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0053] Embodiment Twelve pairs of microsatellite primers for Chrysanthemum chrysophyllum microsatellite primers, amplification and sequencing methods include the following steps:
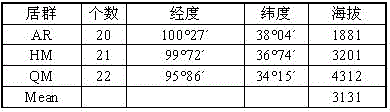
[0054] (1) Genomic DNA of the C. chrysogenum population of 63 individuals of the following three flora was extracted using the improved CTAB method (see Table 1).
[0055] Table 1 Chrysanthemum viridans ( F. luteovirens ) sample collection information
[0056]
[0057] Specific steps are as follows:
[0058] ① Weigh 0.5 g of the internal tissue at the junction of the fruit body stipe and the cap after drying in an oven at 45°C, add liquid nitrogen to the mortar for precooling, add 0.2 g of PVP powder, and put the material into the liquid Grind evenly in nitrogen until all are ground to powder;
[0059] ② Transfer to a 1.5 ml centrifuge tube, add 600 μL BI solution preheated at 65°C, shake in a water bath at 65°C for 10 minutes, centrifuge at 10,000 rpm at 4°C for 10 minutes, and discard the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com