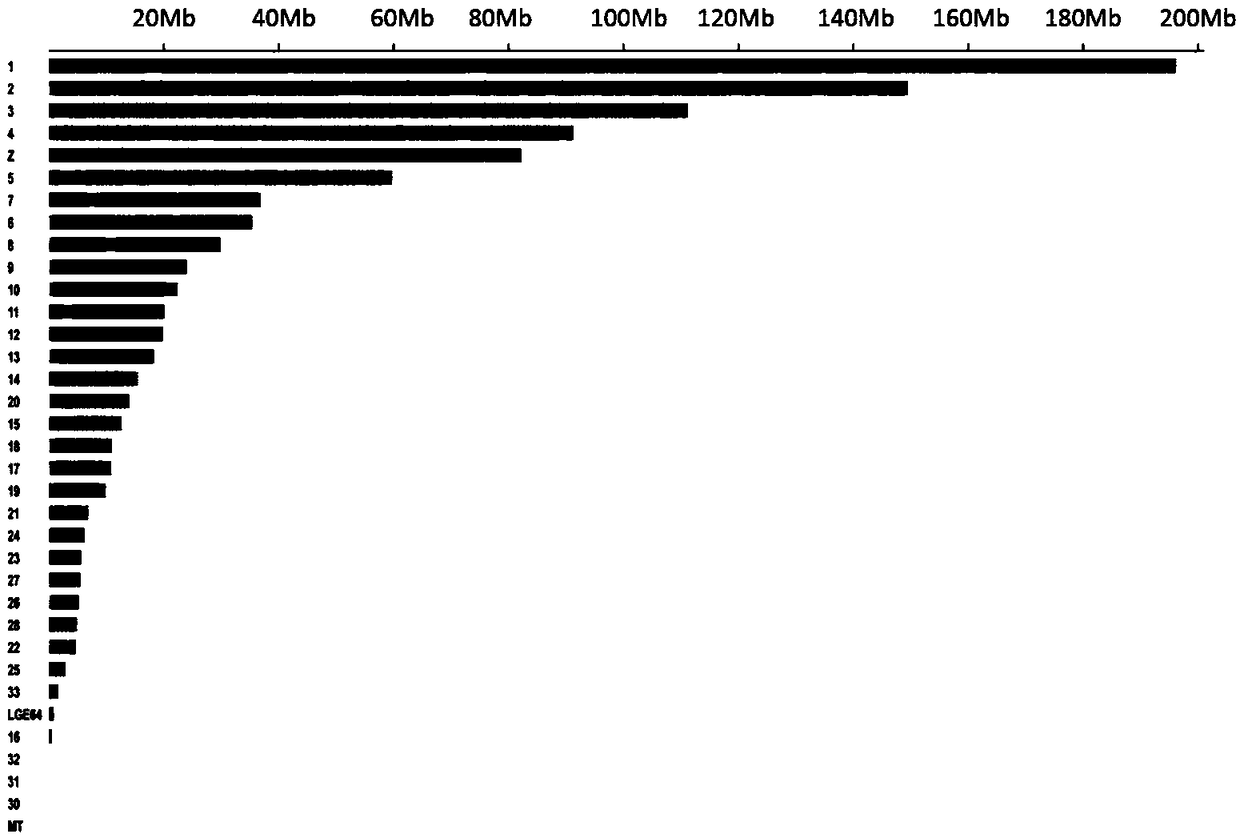
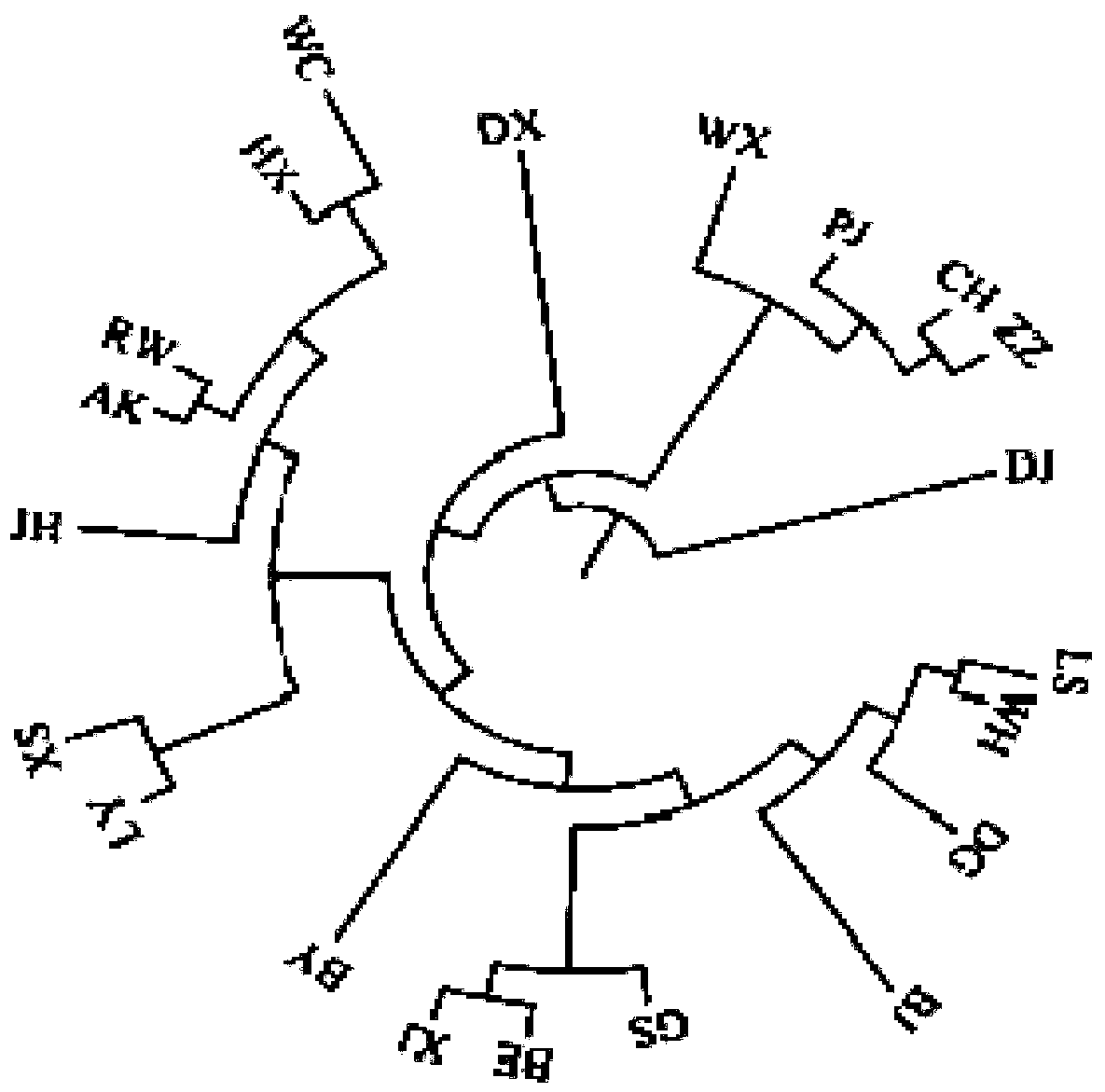
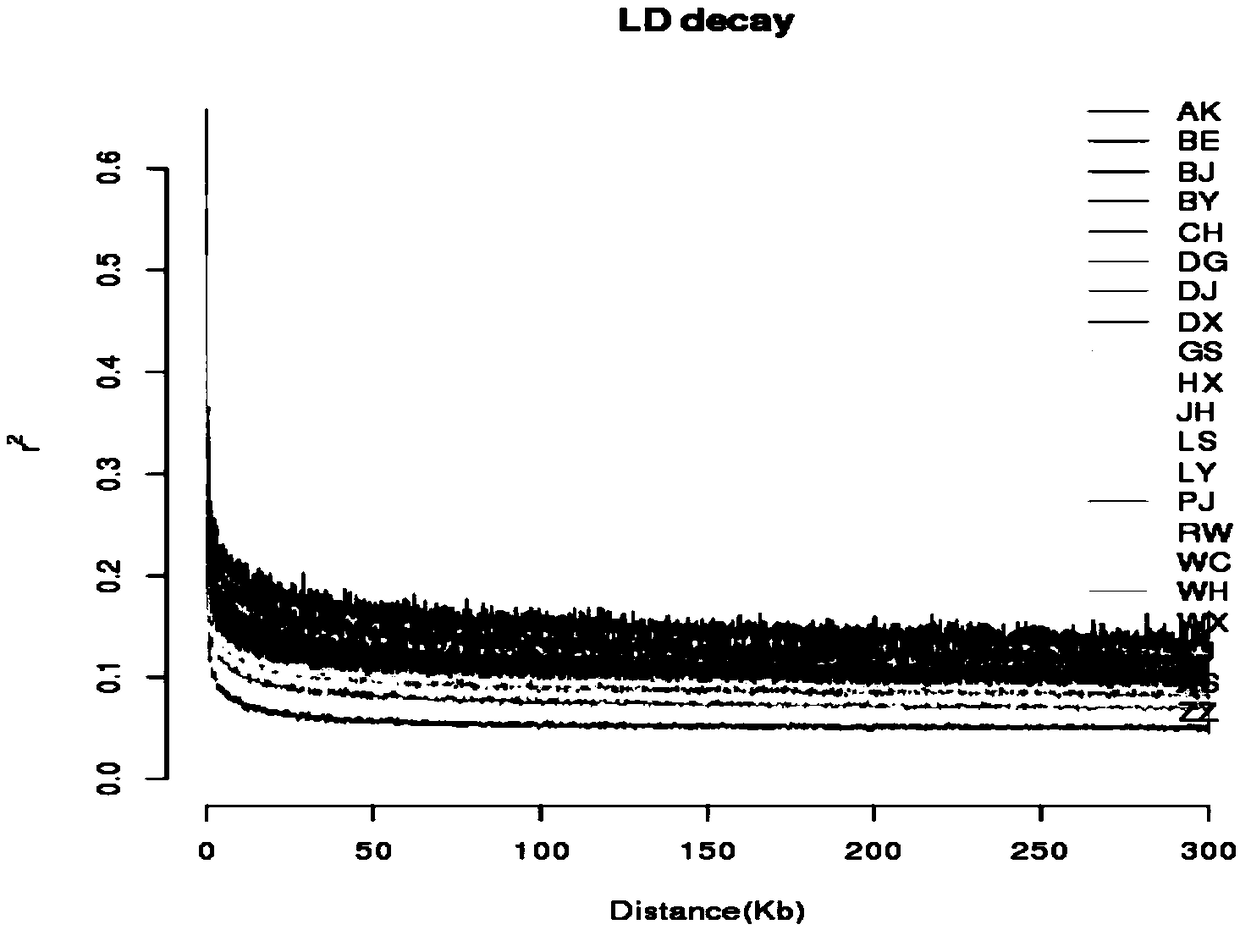
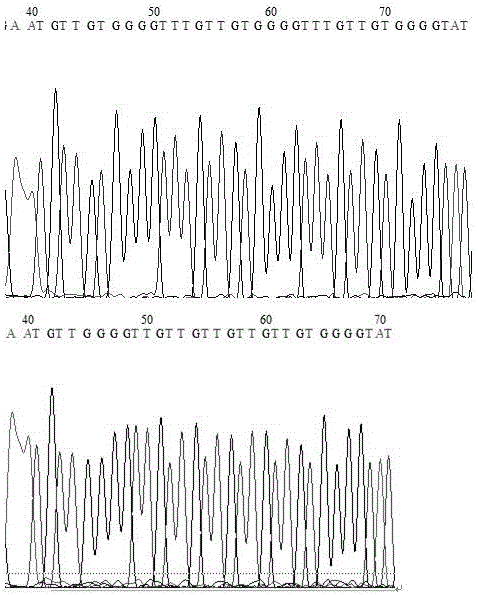
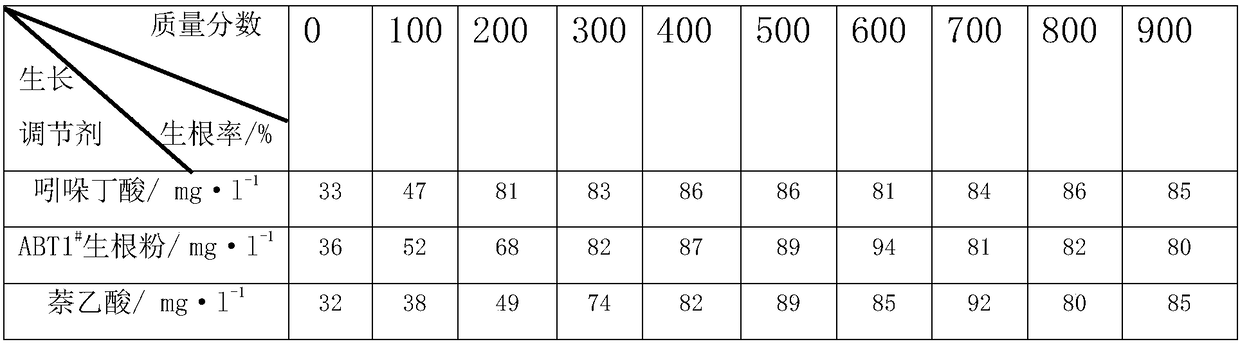
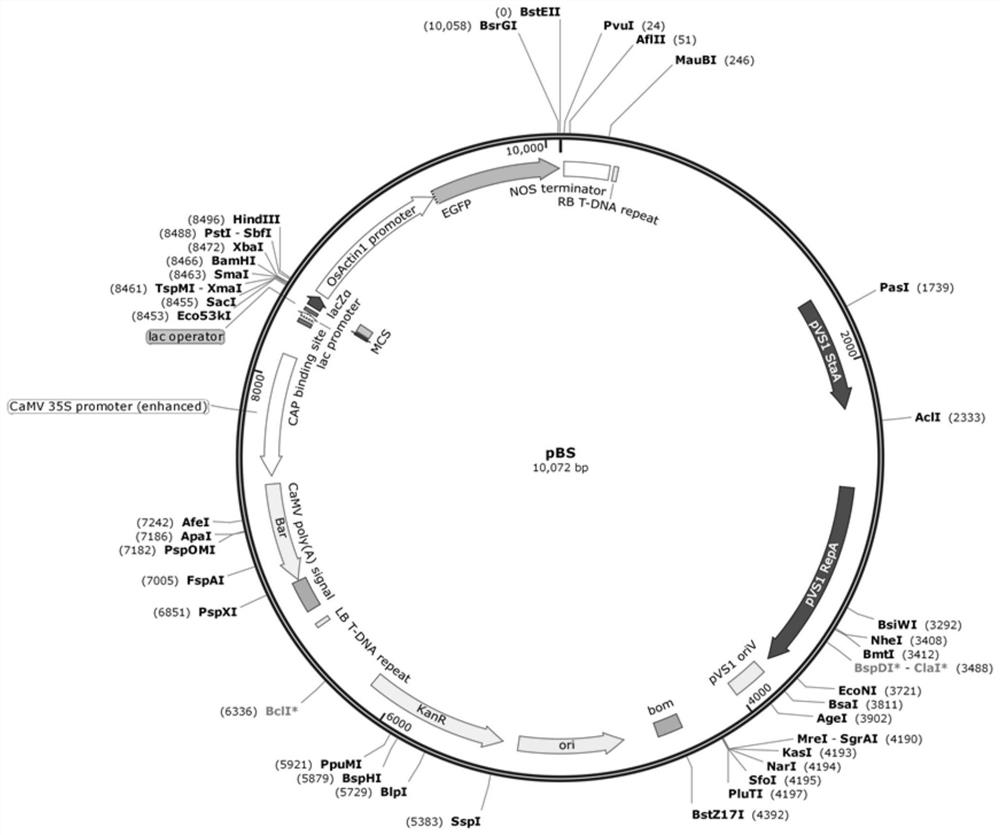
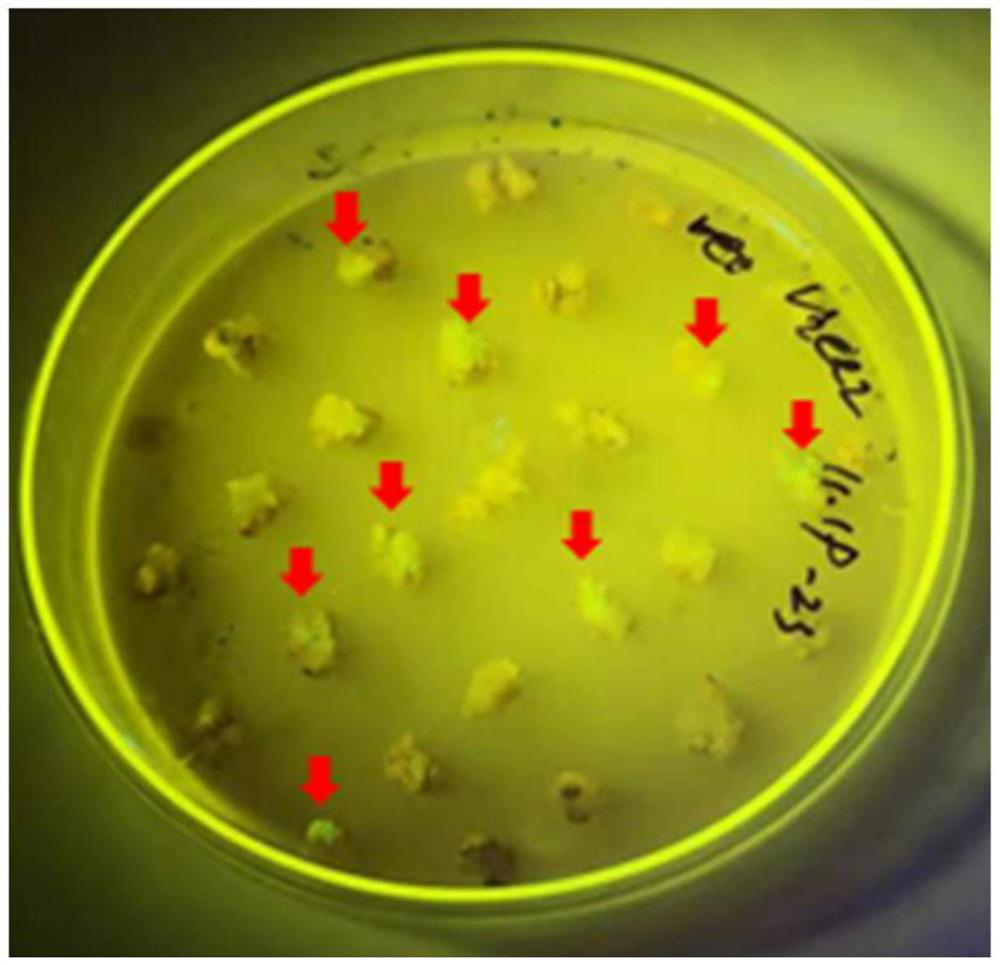
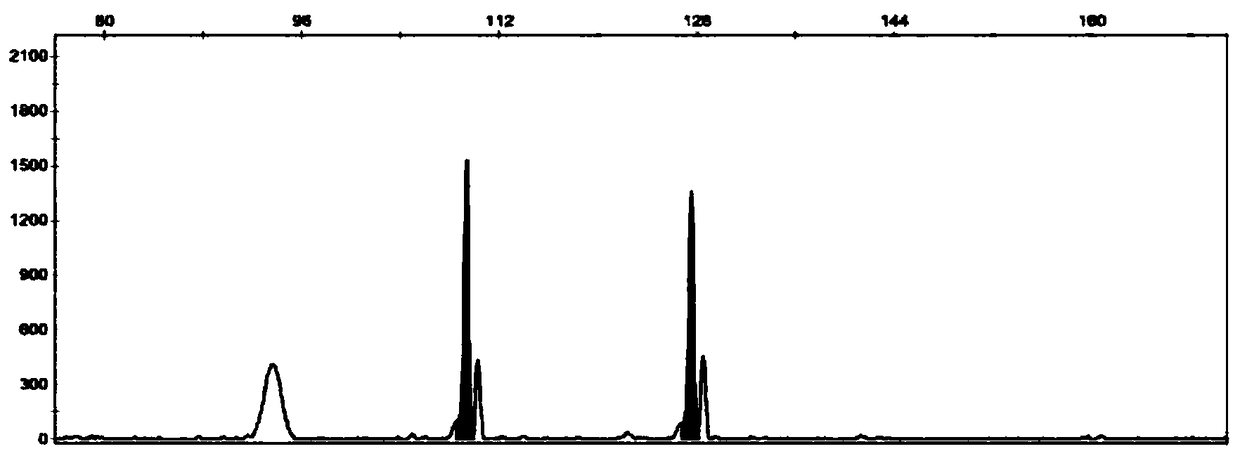
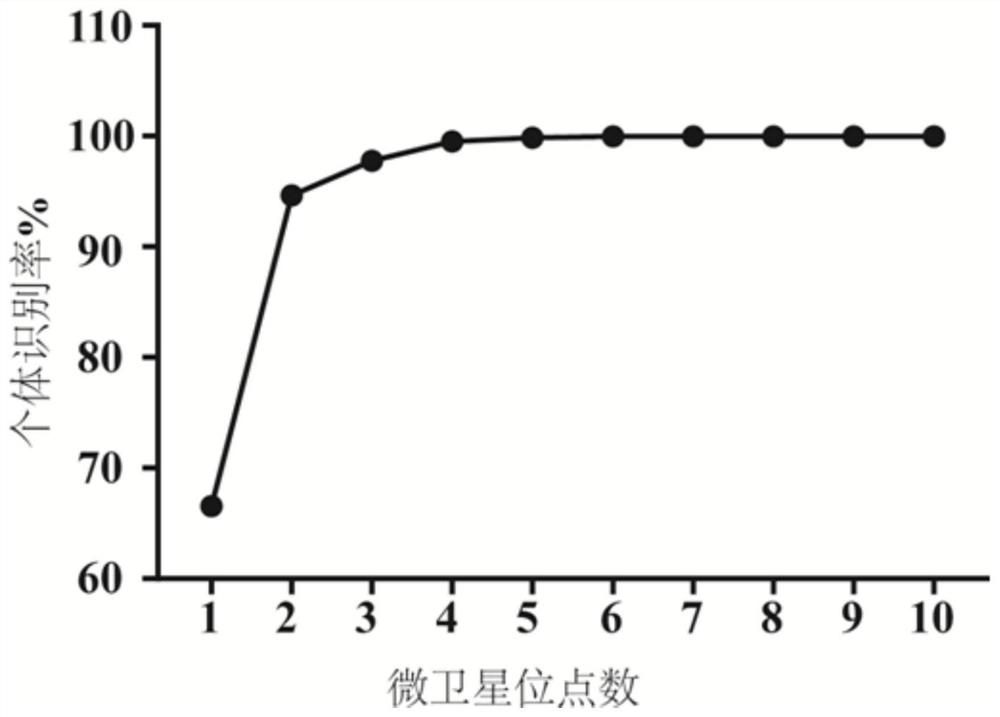
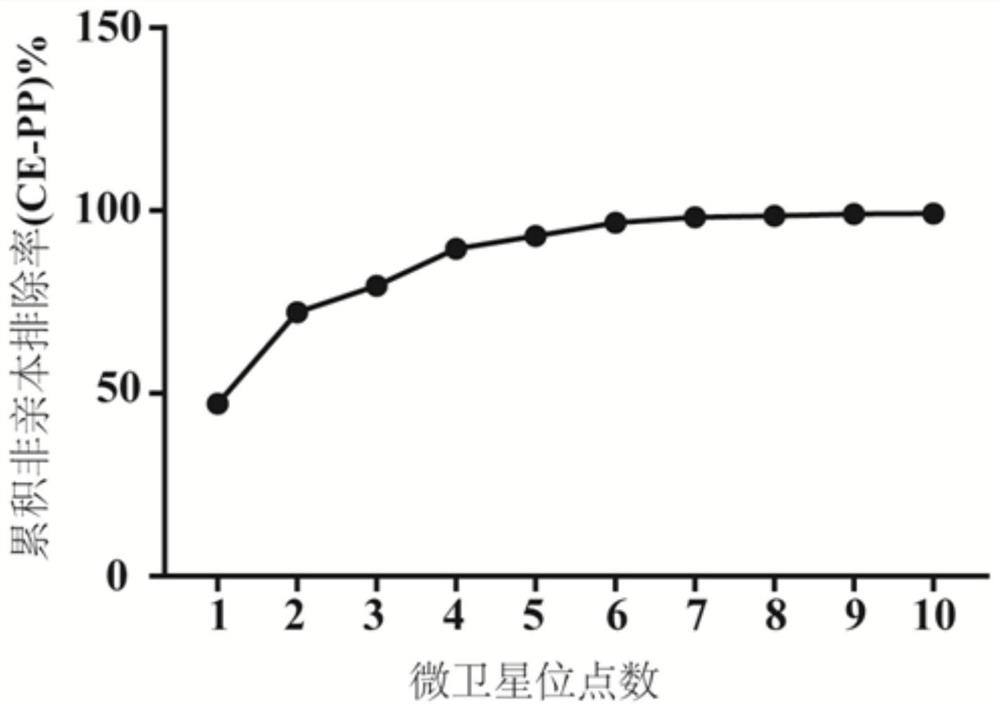
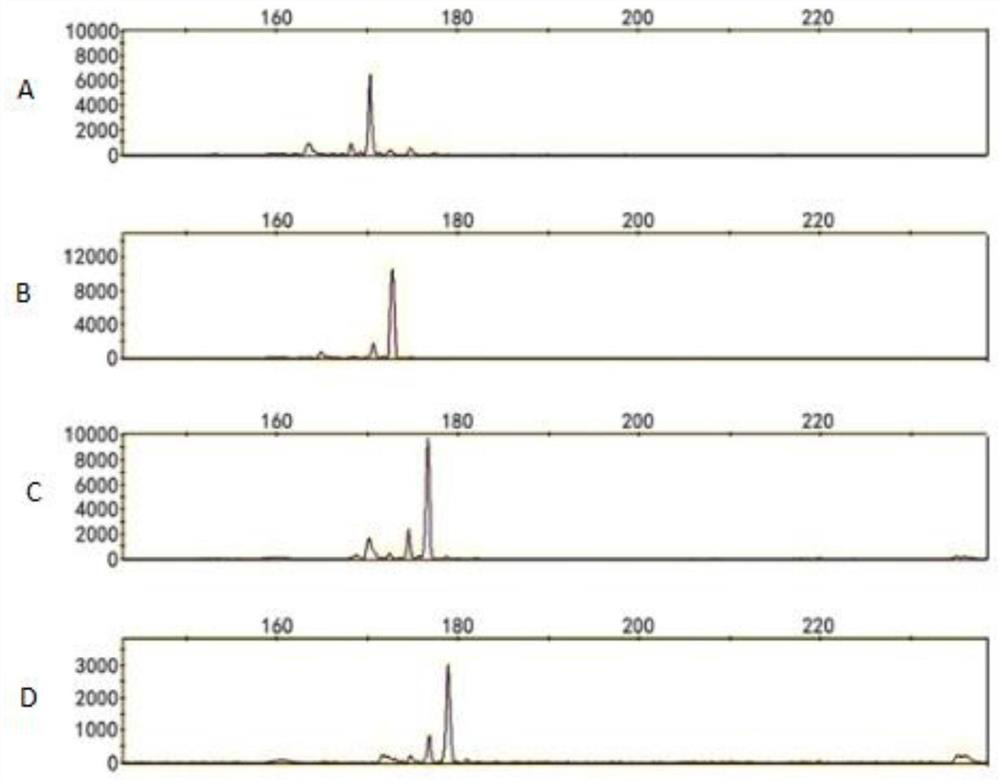
Patents
Literature
38 results about "Genetic differentiation" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Genetic differentiation. [jə¦ned·ik ‚dif·ə‚ren·chē′ā·shən] (genetics) The accumulation of differences in allelic frequencies between completely or partially isolated populations due to evolutionary forces such as selection or genetic drift.
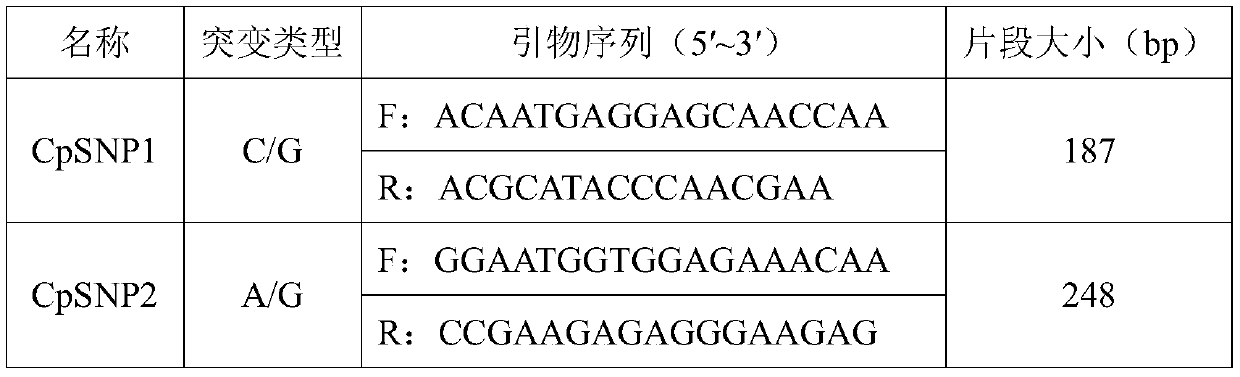
Screening method and application of Dongxiang blue-eggshell chicken genome SNP molecular marker
PendingCN109295238AComprehensive and effective germplasm analysisPromote development and utilizationMicrobiological testing/measurementEggshellScreening method
Relating to the field of genetics, the invention specifically provides a screening method and application of a Dongxiang blue-eggshell chicken genome SNP molecular marker. The screening method includes: conducting high-throughput sequencing detection, then calculating the genetic statistics, then analyzing the genetic difference between Dongxiang blue-eggshell chickens and other chicken breed groups, and finally screening out the Dongxiang blue-eggshell chicken SNP molecular marker. The screening method has the technical effects of efficient and rapid screening of Dongxiang blue-eggshell chicken molecular marker, the selected gene of the screened Dongxiang blue-eggshell chicken serves as the molecular marker for identifying the Dongxiang blue-eggshell chicken germplasm, has the characteristics of low cost, abundant data and high detection precision, alleviates the technical problems of high genetic differentiation research cost and low accuracy caused by lack of efficient molecular biological method and genetic marker in current Dongxiang blue-eggshell chicken genetic differentiation identification. And application of the Dongxiang blue-eggshell chicken genome SNP molecular markeralleviates the technical problem of lack of efficient molecular biological marker in the genetic differentiation research of Dongxiang blue-eggshell chicken.
Owner:JIANGSU INST OF POULTRY SCI
Method for designing, amplifying and sequencing twelve pairs of floccularia luteovirens microsatellite primers
InactiveCN105969862AEliminate distractionsEliminate cumbersome stepsMicrobiological testing/measurementDNA preparationNucleotide diversityFloccularia luteovirens
The invention relates to a method for designing, amplifying and sequencing twelve pairs of floccularia luteovirens microsatellite primers. The method comprises the following steps: (1) extracting genome DNA of three floccularia luteovirens populations among which the geographic interval is more than 300km by using an improved CTAB method; (2) randomly selecting an individual genome DNA respectively from the three populations, mixing, detecting the quality of total DNA, preparing a gene library, and performing Illumina HiSeq<TM>2500 sequencing after the gene library is qualified in examination in depot; (3) splicing sequenced data, detecting simple sequence repeats (SSR) in the total DNA sequence by using SR search software, and performing primer design by applying primer3; (4) preparing an SSR primer having an annealing temperature of 50-60 DEG C by adopting a temperature gradient method; (5) respectively performing PCR amplification on the genome DNA of the three floccularia luteovirens populations, and sequencing and verifying to obtain 12 pairs of primers with polymorphism; and (6) calculating the number N of allelic genes, haplotype diversity H<d> genetic differentiation coefficient F<ST>, nucleotide diversity P, G<ST> and the value of pi. The method is beneficial to large-scale research.
Owner:CHINA ACAD OF SCI NORTHWEST HIGHLAND BIOLOGY INST
Auxiliary screening method for indica rice and japonica rice, and special-purpose primer for the same
InactiveCN101215608AUnderstanding Degrees of Genetic DifferentiationOptimize hybrid mixMicrobiological testing/measurementDNA/RNA fragmentationOryzaGermplasm
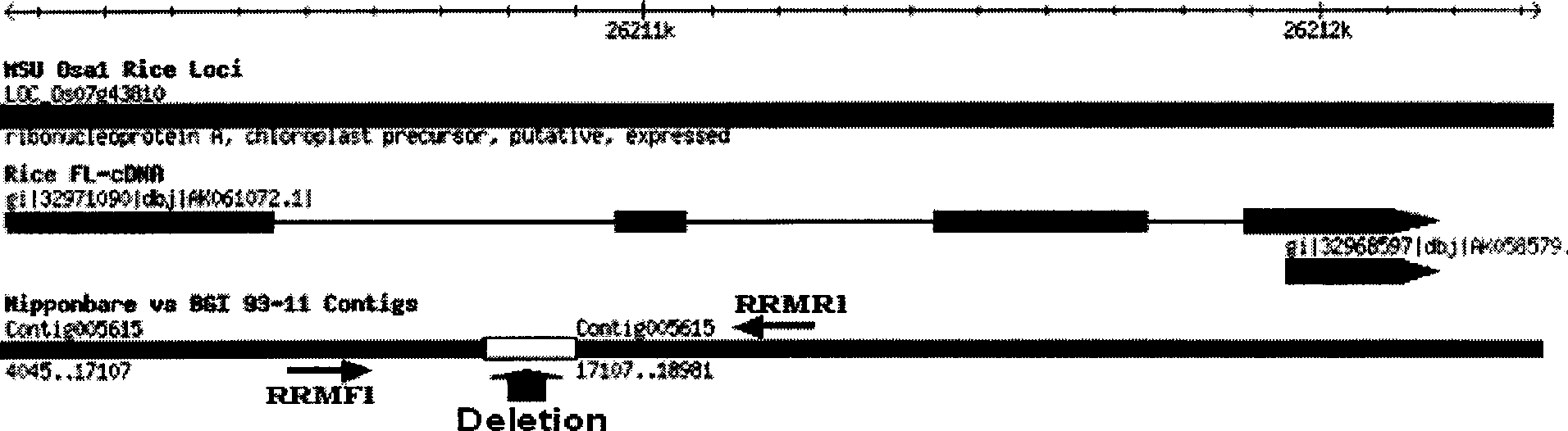
The invention discloses a process of auxiliary screening hsien rice and japonica rice. The process for auxiliary screening hsien rice and japonica rice which is provided by the invention is used to detect whether rice losses ribonucleotide sequence of 122836-124566 from 5' end in AC132003, the detected rice is candidate hsien rice if lost, or the detected rice is candidate japonica rice if not lost. The process for auxiliary screening rice subspecies which is provided by the invention can be used to evaluate hsien rice and japonica rice breeds or local breed in early period, to find out genetic differentiation of hsien rice and japonica rice, and plays important guiding roles for using heterosis of hsien rice and japonica rice and cultivating high-yield and high grade hybridization rice, thereby optimizing hybridized combination, accelerating breeding speed, reducing breeding cost, and having the advantages of simple operation, low cost and short cycle, which is suitable to be popularized and applied, and provides a fast choose method for the evaluation of rice subspecies germplasm.
Owner:CHINA AGRI UNIV
Persimmon tree colletotrichum gloeosporioide SSR (Simple Sequence Repeat) primer pair developed based on sibling species genome and application of primer pair
ActiveCN106701943AEffective prevention and controlMicrobiological testing/measurementMicroorganism based processesConsanguinityGenome
The invention belongs to the technical field of biologics, and in particular discloses a persimmon tree colletotrichum gloeosporioide SSR (Simple Sequence Repeat) primer pair developed based on sibling species genome and application of the primer pair. According to the persimmon tree colletotrichum gloeosporioide SSR primer pair, 30 pairs of SSR primers are developed by finding SSR by using MISA software on the basis of the genome of a sibling species, namely, colletotrichum gloeosporioides, of persimmon tree colletotrichum gloeosporioides, and 15 pairs of polymorphism SSR primers are obtained by screening 6 different persimmon tree colletotrichum gloeosporioides; the persimmon tree colletotrichum gloeosporioide SSR primer pairs provided by the invention are novel markers which exist stably, can be used for analyzing genetic diversity and genetic differentiation on the persimmon tree colletotrichum gloeosporioides, can be also used for identifying varieties of the persimmon tree colletotrichum gloeosporioides and studying ties of consanguinity, and have very important significances in effective prevention and control on the persimmon tree colletotrichum gloeosporioides.
Owner:河南省林业科学研究院
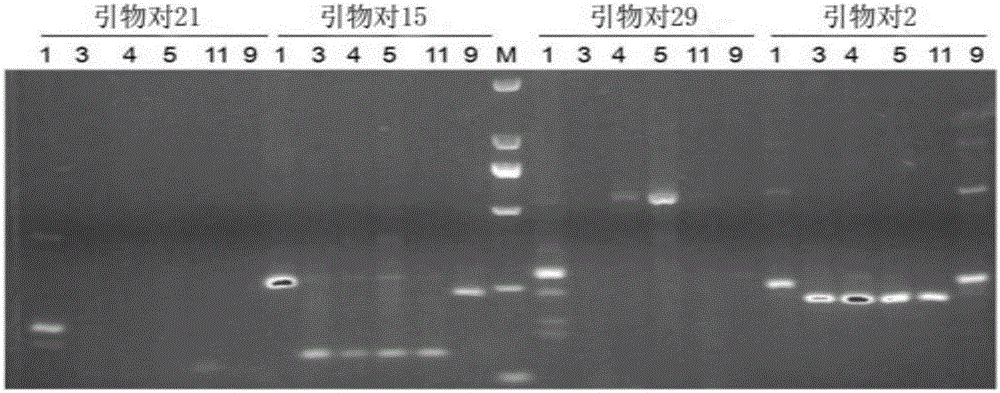
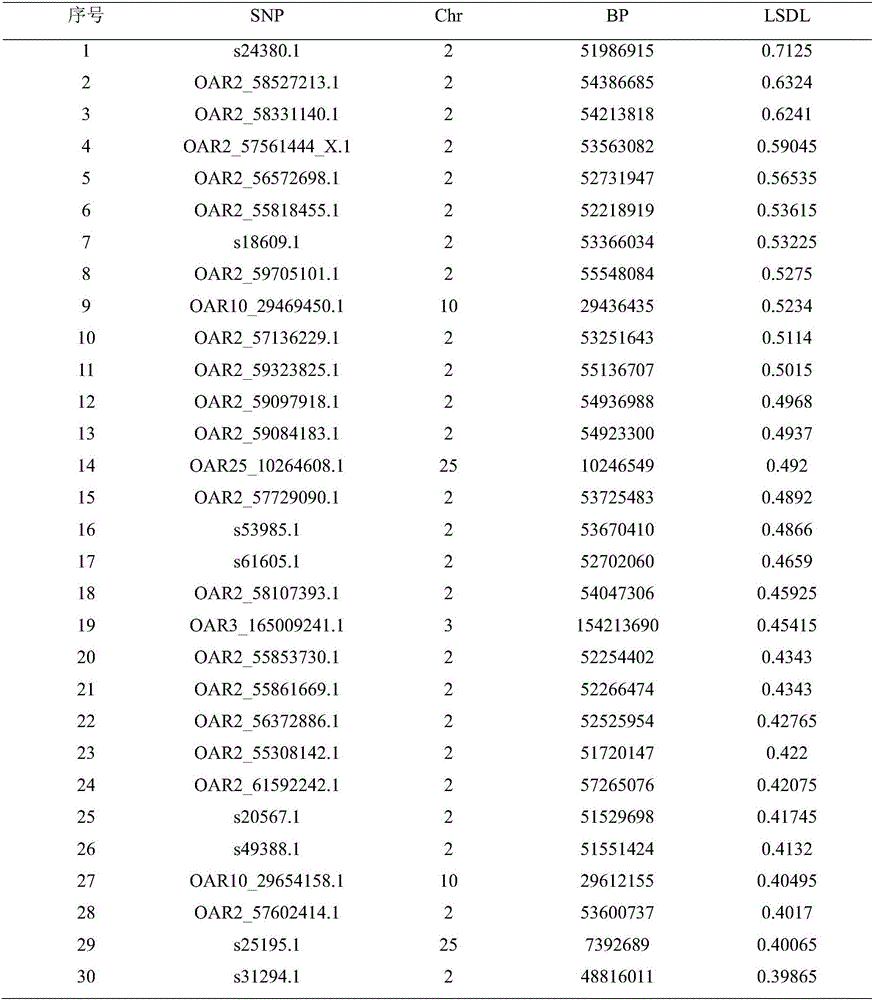
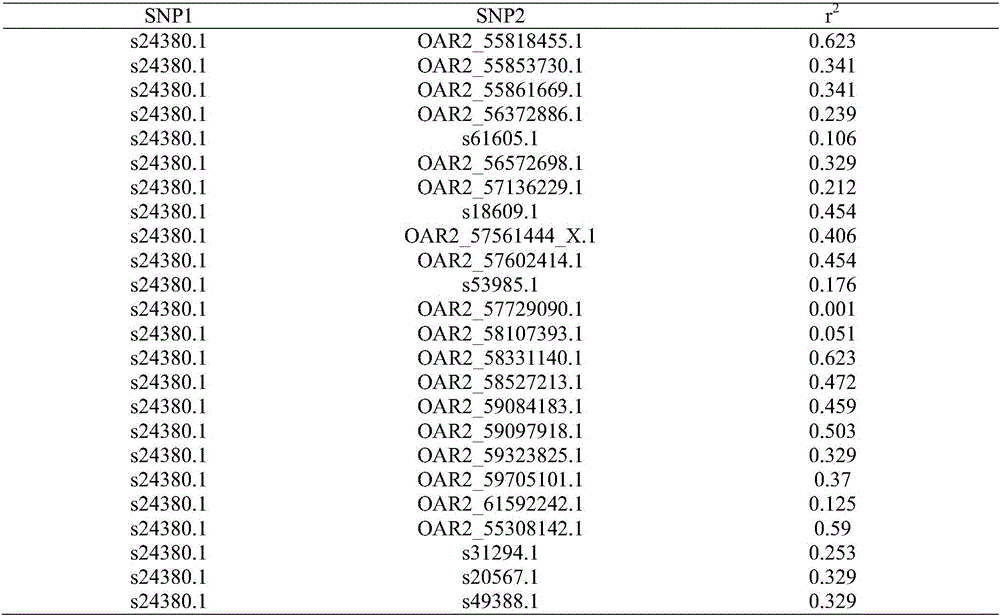
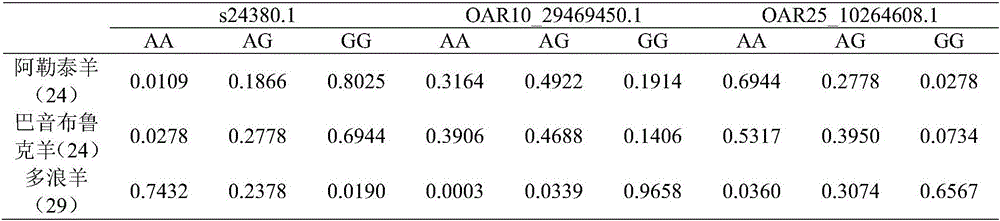
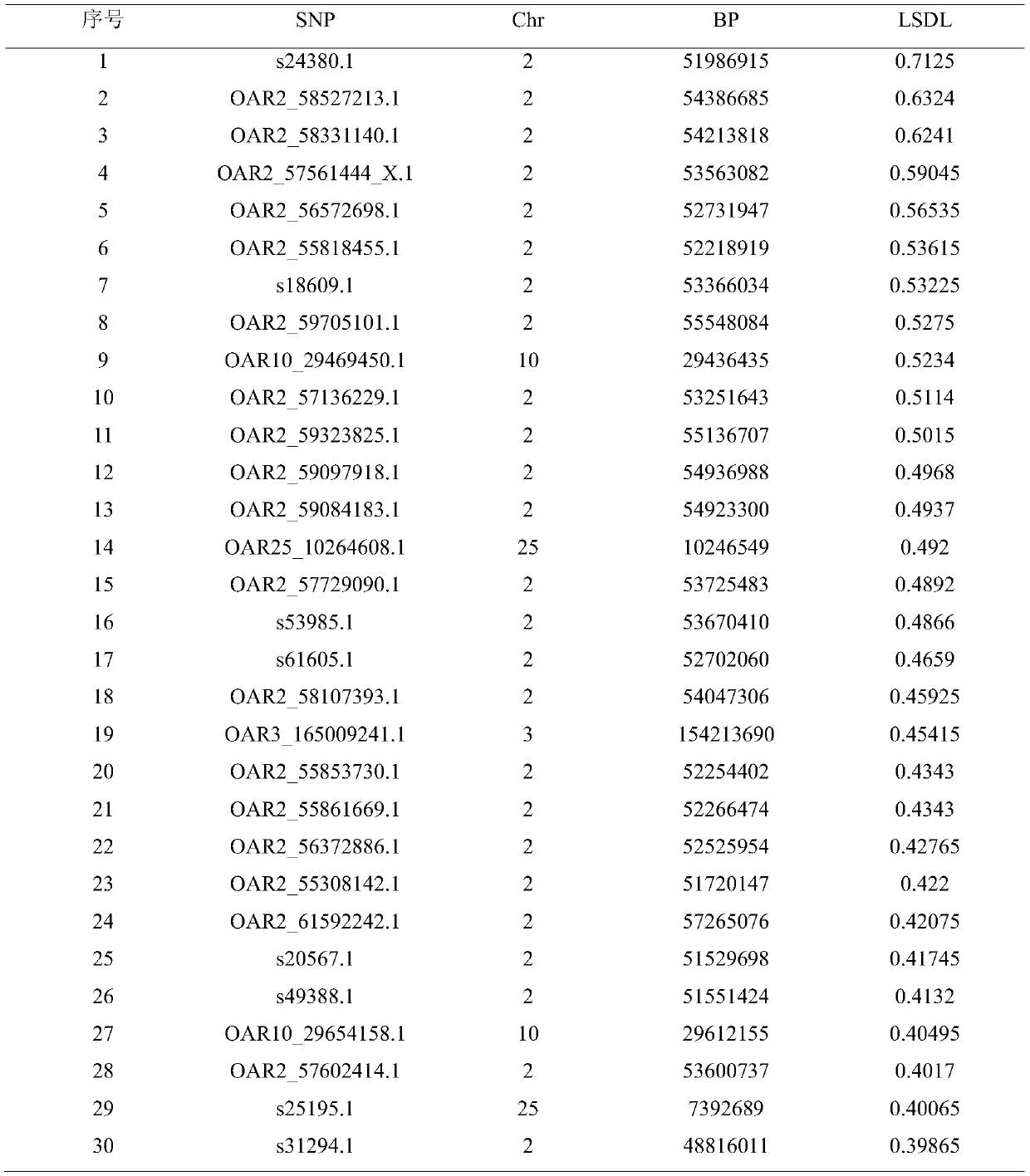
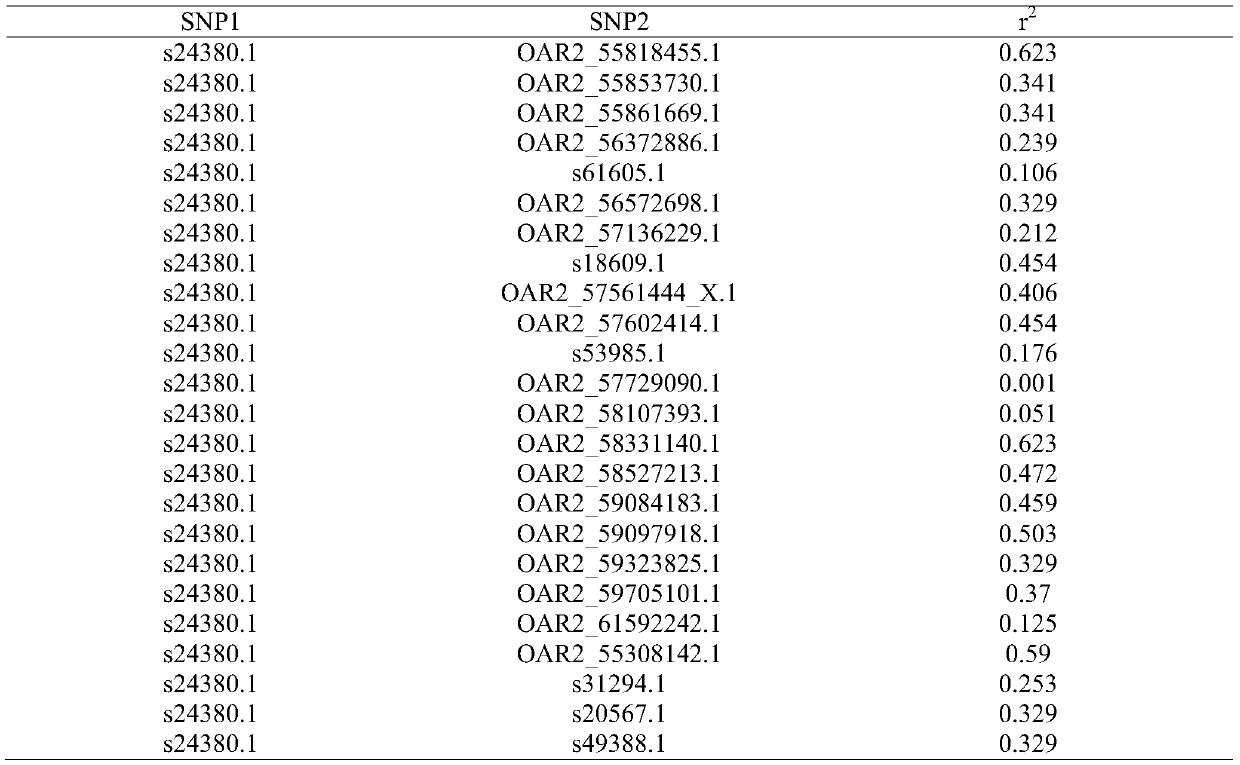
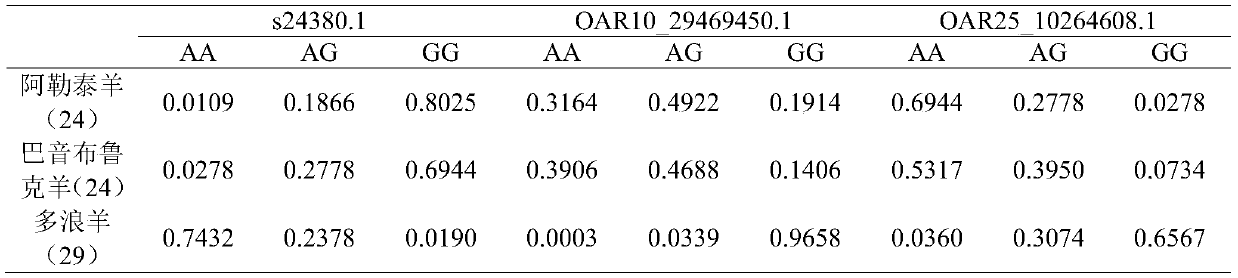
Duolang sheep SNP (single-nucleotide polymorphism) marker, and screening method and application thereof
ActiveCN106011259AImprove accuracyOvercoming the pitfalls of phenotypingMicrobiological testing/measurementDNA/RNA fragmentationGenetic linkage disequilibriumScreening method
The invention discloses a Duolang sheep SNP (single-nucleotide polymorphism) marker, and a screening method and application thereof, belonging to the field of animal molecular markers. A genetic differentiation coefficient method is utilized to perform screening on a commercialized sheep genome SNP chip and further remove high-linkage-disequilibrium SNP markers (r<2>>0.2) through linkage disequilibrium analysis, thereby obtaining the Duolang sheep SNP marker. The Duolang sheep SNP marker can be applied to identification of Duolang sheep species or animal products thereof on the premise of no obvious linkage disequilibrium among markers; and therefore, when the Duolang sheep SNP marker disclosed by the invention is used for identifying the Duolang sheep species or animal products thereof, no interference among sites exists, thereby greatly enhancing the Duolang sheep species identification accuracy and effectively lowering the identification cost.
Owner:新疆畜牧科学院生物技术研究所
Method for predicting percentage of fructification of indica-japonica hybrid F1 by using InDel molecular markers
InactiveCN103798127AGuaranteed outputMicrobiological testing/measurementPlant genotype modificationBiotechnologyMicrobiology
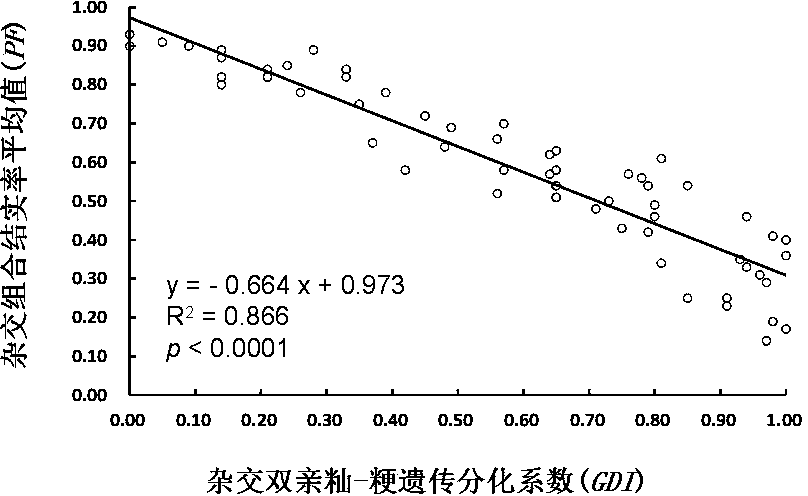
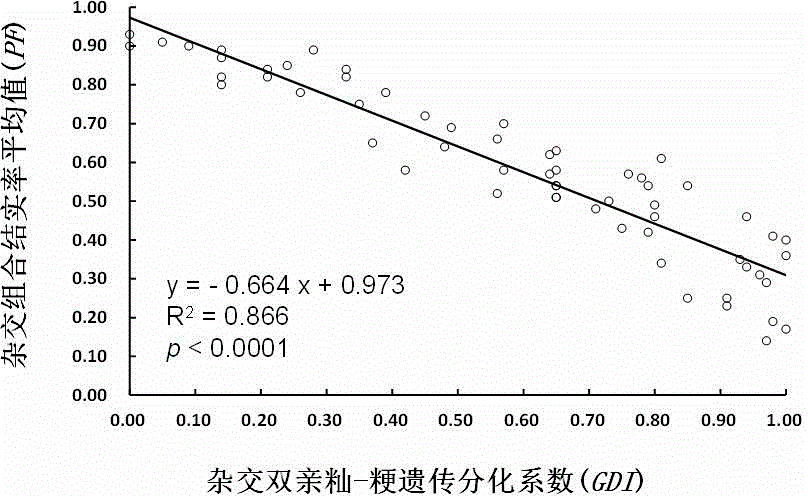
The invention belongs to the technical field of molecular mark assisted breeding, and particularly discloses a method for detecting genetic differentiation level of indica-japonica hybridizing parents and predicting the percentage of fructification of hybrid F1 by using InDel molecular markers. The hybridizing parents can be purposefully selected and the hybrid F1 with high percentage of fructification is prepared by detecting the genetic differentiation level of the indica-japonica hybridizing parents. The indica-japonica genetic differentiation coefficients (GDI) of indica-japonica hybridizing parents are determined by using 40 InDel molecular markers and a built calculation method, the correlation between the indica-japonica hybridizing parents GDI and a really measured mean value of the percentage of fructification (PF) of the hybrid F1 is really analyzed, so as to form a method for analyzing parents GDI to predict the percentage of fructification of the indica-japonica hybrid F1 by the molecular markers. The indica-japonica genetic differentiation coefficient detection on the parents can be carried out before hybridization, so as to predict the percentage of fructification of the hybrid F1. Thus, the indica-japonica hybridizing parents can be effectively selected.
Owner:FUDAN UNIV
Method for assisting sifting motion of nonglutinous rice and non glutinous rice
InactiveCN101220394AUnderstanding Degrees of Genetic DifferentiationOptimize hybrid mixMicrobiological testing/measurementDNA/RNA fragmentationOryzaGermplasm
The invention discloses an assisted screening method of indica type rice and japonica rice. The assisted screening method of indica type rice and japonica rice which is provided by the invention is to detect whether the 71046 to 72062 nucleotide sequence from the 5' tail end of GenBank accession AP003771 is deleted in rice to be measured, if deleted, the rice to be measured is the candidate indica type rice, if not deleted, the rice to be measured is the candidate japonica rice. The assisted screening method of indica type rice and japonica rice of the invention can be used for identifying the variety and the local races of the indica type rice and the japonica rice at an early stage and knowing the genetic differentiation degree of the indica type rice and the japonica rice, thus the invention has important guide function for utilizing the hybrid advantages of the indica type rice and the japonica rice and cultivating the high-yield and high-quality hybrid rice so as to optimize hybrid combination, accelerate breeding speed and reduce breeding cost; the invention has the advantages of simple operation, low cost and short period, which is applicable to popularization and application and provides a fast choosing method for the identification of the germplasm of the subspecies of the indica and japonica of the rice.
Owner:CHINA AGRI UNIV
Peony genome DNA molecule marking method
InactiveCN103820546AEasy to operateLow costMicrobiological testing/measurementDNA/RNA fragmentationElectrophoresesEnzyme digestion
A peony genome DNA molecule marking method is as follows: extracting a peony genomic DNA for enzyme digestion, connecting an enzyme digestion product with a joint DNA; performing PCR amplification of a connection product and a pre amplified primer to obtain a pre amplified product; taking the pre amplified product for PCR amplification with iPBS primer which is selectively amplified by use of an incision enzyme joint to obtain a PCR selectively-amplified product; then taking the PCR selectively-amplified product to add into a dyeing liquid, applying a sample for electrophoresis, after the electrophoresis, silver staining, observing, photographing and analyzing results. The peony genome DNA molecule marking method is a molecule marking technology based on the PCR and enzyme digestion, and has the advantages of simple operation, low cost and strong applicability; designed for the peony genome study technology, a new molecule marking system based on the retrotransposon is developed, and provides a new technological means for researches on genetic differentiation and evolution of germplasm resources of peony.
Owner:HENAN UNIV OF SCI & TECH
Method for screening manganese/cadmium absorption efficient ecological phytolacca americana
InactiveCN107475384AComparableEasy to operateMicrobiological testing/measurementSystems biologyManganeseScreening method
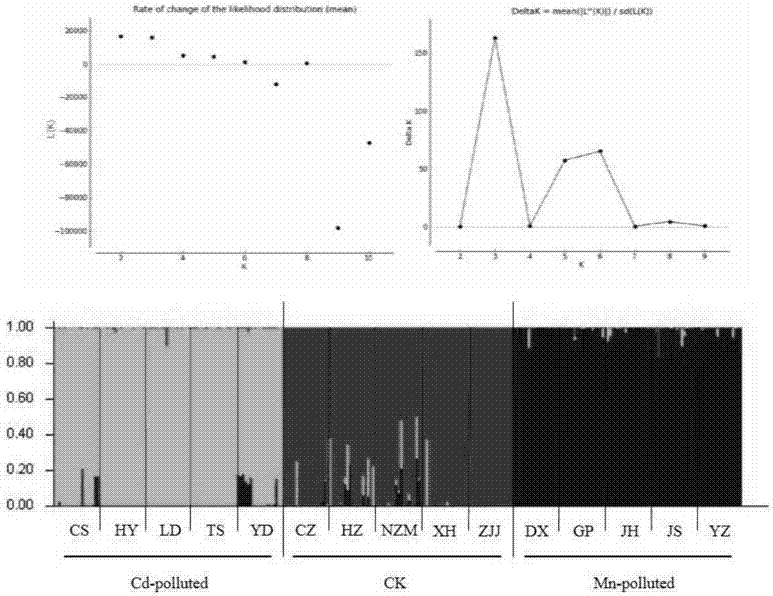
The invention relates to a plant restoration technology, and particularly discloses a method for screening manganese / cadmium absorption efficient ecological phytolacca americana. The method comprises the following steps: collecting in the wild leaves and seeds of phytolacca americana populations in a plurality of contaminated regions and uncontaminated regions in China and soil in the vicinity; detecting the manganese / cadmium content of the soil in the vicinity of the phytolacca americana populations in the regions, and selecting phytolacca americana populations in highly manganese contaminated, highly cadmium contaminated and uncontaminated regions respectively; performing AFLP (amplified fragment length polymorphism) molecular marker detection on the leaves of the selected phytolacca americana populations, and analyzing genetic structures thereof; selecting bred seedlings of the phytolacca americana populations to which different genetic differentiation occurs, culturing the seedlings for 50 days with manganese, cadmium and normal Hoagland solutions, and screening the manganese / cadmium absorption efficient ecological phytolacca americana by adopting the plant heights, fresh weights and leaf manganese / cadmium content of the phytolacca americana as indexes. According to the method, a manganese / cadmium absorption efficient ecological hyperaccumulator is used for treating manganese / cadmium-contaminated soil, and the heavy metal accumulation and restoration efficiency of the hyperaccumulator are improved.
Owner:NANJING AGRICULTURAL UNIVERSITY
Screening method and application of Bian chicken molecular marker
ActiveCN109295239AImprove detection accuracyLow costMicrobiological testing/measurementBiotechnologyScreening method
Relating to the field of genetics, the invention provides a screening method and application of a Bian chicken molecular marker. The screening method of a Bian chicken molecular marker includes: conducting high-throughput sequencing detection on SNP marker, calculating genetic statistics, analyzing and comparing the genetic difference of Bianji group and other chicken breed groups, and screening out a Bian chicken molecular marker. The method provided by the invention alleviates the technical problems of high Bian chicken genetic differentiation research cost and low accuracy caused by lack ofefficient molecular biological method in existing Bian chicken breed conservation effect evaluation process, and reaches the technical effect of efficient and rapid screening of a molecular marker for Bian chicken breed conservation effect evaluation. The invention also provides a screening method of the Bian chicken molecular marker and application of the screened molecular marker, and alleviates the technical problems of lack of efficient molecular biological marker and screening method in Bian chicken research.
Owner:JIANGSU INST OF POULTRY SCI
Method for detecting number of littermate and characters of sow
InactiveCN101956009AUnderstanding Degrees of Genetic DifferentiationImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationDNA fragmentationHaplotype
The invention discloses a method for detecting number of littermates and characters of a sow, which comprises the steps of: 1, detecting whether the 71th nucleotide from 5' end of a DNA segment shown in SEQ ID NO:1 in a genome of a sow to be detected is G or A; detecting whether the 81th nucleotide from 5' end of a DNA segment shown in SEQ ID NO:2 in the genome of the sow to be detected is C or A; and determining the haplotype of the sow to be detected, determining the number of the littermates and the characters of the sow to be detected according to the haplotype. The method can be used for detecting reproduction characters of the sow in a development early stage and knowing the genetic differentiation degree of the sow, provides a useful molecular marker for genotyping, screening and auxiliary breeding of different haplotypes of the sow, and has important meanings on improving the number of the littermates of the sow. Therefore, the method is beneficial to the quickening of the breeding speed and the reducing of the breeding cost, has the advantages of low cost, simple operation and short period, and is suitable for popularization and application.
Owner:CHINA AGRI UNIV
Method for detecting single nucleotide polymorphism of genes related to yean trait of milk goat
InactiveCN107217091ASpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationGenomicsNonsynonymous substitution
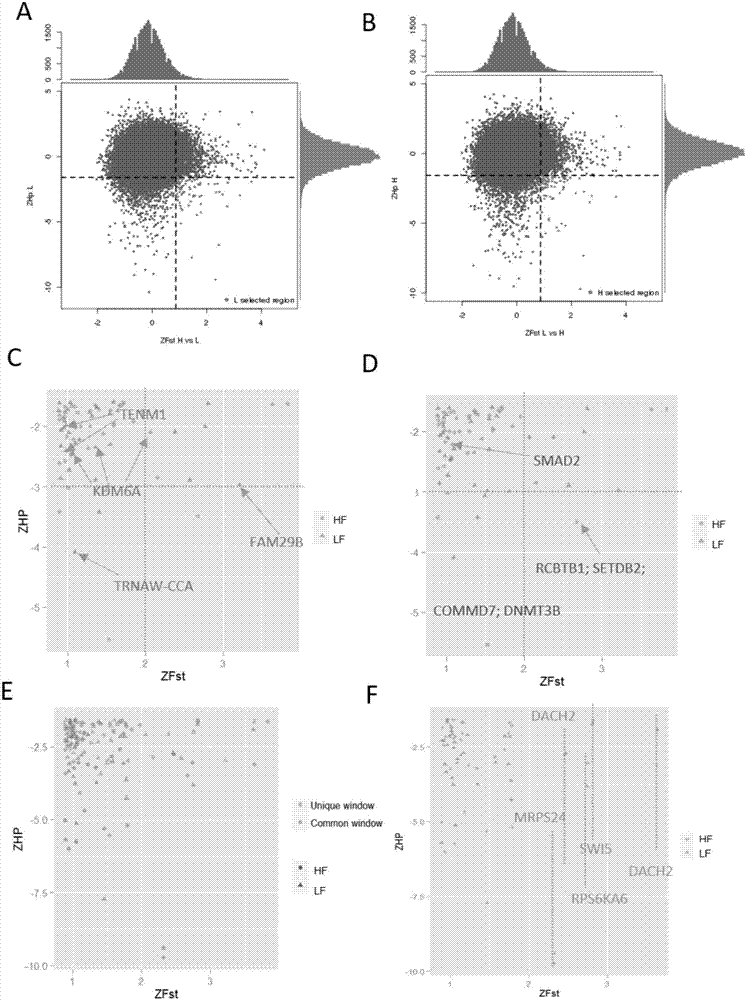
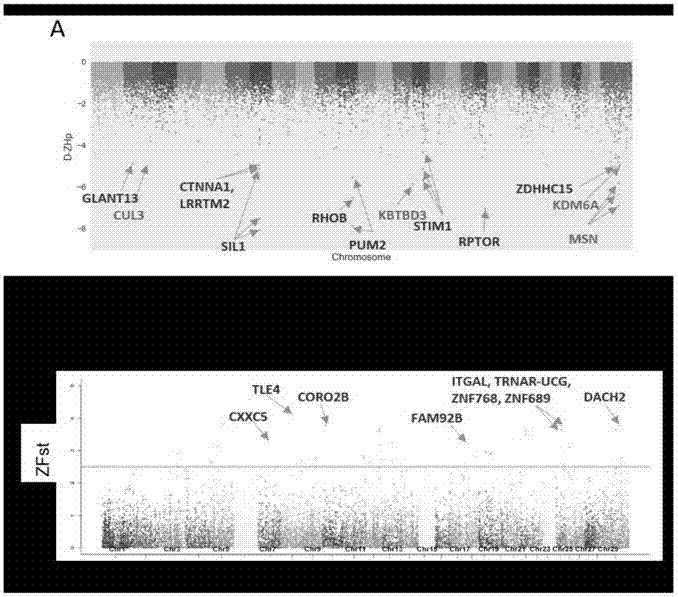
The invention relates to the technical fields of high-throughput sequencing, genomics and bioinformatics, in particular to a detection method for analyzing single nucleotide polymorphism of genes related to yean trait of a milk goat and candidate genes by adopting a selective elimination analysis method on the basis of the single nucleotide polymorphism of the milk goat. The method comprises the following steps of 1) comparing data passing through the high-throughput sequencing to a reference genome, then annotating SNP (single nucleotide polymorphism) sites of the milk goat by utilization of free ANNOVAR software; 2) evaluating a sequencing sequence of two groups with different yean properties by utilization of a genetic differentiation coefficient (Fst) and heterozygosity scores (Hp) so as to screen out SNPs of each sliding window; 3) screening out homozygous SNPs from a low-yield group and a high-yield group, and authenticating single-base nonsense substitution and nonsynonymous substitution specifically existed in the two groups. The method disclosed by the invention has the advantages that the structure and function of protein may be affected due to the fact that the translation and amino acid sequences of the protein are directly affected through two variations, namely the single-base nonsense substitution and the nonsynonymous substitution.
Owner:QINGDAO AGRI UNIV
Method for assistantly screening indica-type rice and japonica rice, and special primer therefor
InactiveCN101487055AUnderstanding Degrees of Genetic DifferentiationOptimize hybrid mixMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceFiltration
The invention discloses a filtration assisting method of Indica and Japonica and a specific primer thereof. The filtration assisting method detects whether segments shown in (a) and (b) are deleted in a protein gene that is shown in sequence 4 of an encoding sequence table in a genome of rice to be detected, and if so, the rice to be detected is Indica candidate, otherwise the rice to be detected is Japonica candidate; wherein, (a) is a DNA segment of 717 to 855 nucleotide from 5'-end in the sequence 3 of the sequence table; and (b) is a DNA segment in a nucleotide sequence that has the homology degree of over 97 percent with sequences that are limited by (a). The filtration assisting method of Indica and Japonica subtypes has important guidance in filtering Indica and Japonica varieties or local varieties in an early development stage, acknowledging the genetic differentiation degree between Indica and Japonica, utilizing the advantages of hybrid varieties Indica and Japonica, and culturing high-yield and high-quality hybrid rice, and can consequently optimize hybrid combinations, quicken breeding speed and lower breeding cost, has the advantages of simple operation, low cost and short period, fits for popularization and application, and provides a fast selection method for the quality identification of rice subtypes Indica and Japonica.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
Method for screening molecular marker of Jinhu black-bone chickens and application of molecular marker
ActiveCN109321665ALow costRich dataMicrobiological testing/measurementScreening methodGenetic divergence
The invention provides a method for screening a molecular marker of Jinhu black-bone chickens and application of the molecular marker and relates to the field of genetics. The method for screening themolecular marker of the Jinhu black-bone chickens comprises the steps of detecting an SNP marker through high-pass sequencing, calculating genetic statistics, and analyzing and comparing genetic differences between Jinhu black-bone chicken colonies and other chicken colonies, thereby screening out the molecular marker of the Jinhu black-bone chickens. The technical problem that the research on genetic differentiation of the Jinhu black-bone chickens is high in cost and low in accuracy due to the fact that a process of evaluation on a Jinhu black-bone chicken breed conservation effect is lackof efficient molecular biology methods at present is solved, and the technical effect of efficiently and rapidly screening out the molecular marker for evaluating the Jinhu black-bone chicken breed conservation effect is achieved. The invention further provides the method for screening the molecular marker of the Jinhu black-bone chickens and application of the screened molecular marker. The technical problem that researches on the Jinhu black-bone chickens are lack of molecular biological markers and screening methods is solved.
Owner:JIANGSU INST OF POULTRY SCI
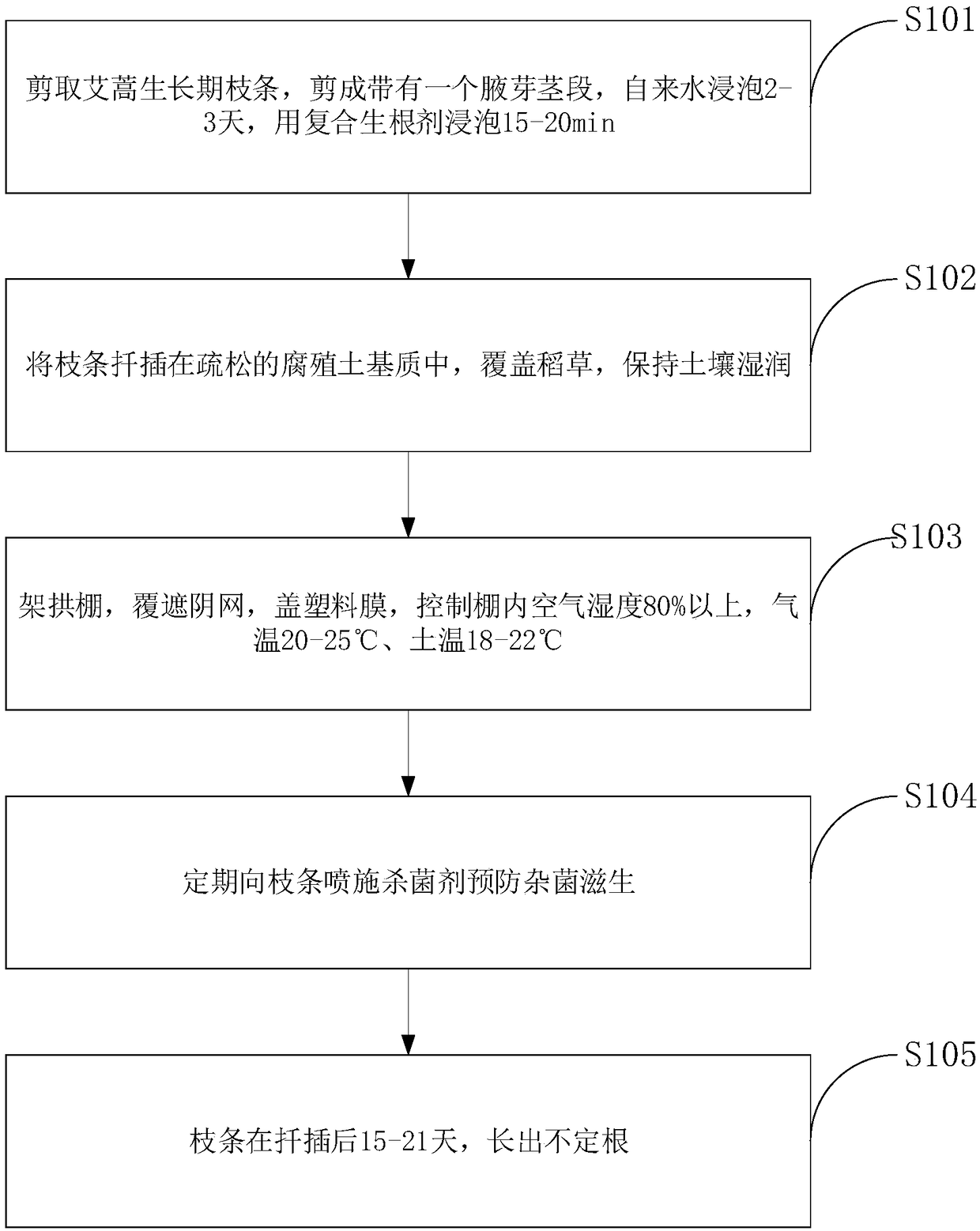
Chinese mugwort cutting propagation method
InactiveCN109197195AExcellent cultivation traitsMaximize yieldVegetative propogationAxillary budShoot
The invention belongs to the technical field of plant vegetative propagation and discloses a Chinese mugwort cutting propagation method. The method includes the following steps that: a Chinese mugwortgrowth-phase shoot is scissored, a stem with axillary buds is obtained, the stem is soaked with tap water and then is soaked with a composite rooting agent; an obtained shoot is inserted into a loosehumus soil matrix and is covered with straw, with the soil kept moist; an arc shed is erected, is covered with a shading net, and is further covered with a plastic film, and air humidity inside the shed is controlled; the shoot is sprayed with a bactericide regularly, so that the growth of bacteria can be prevented; and after the cottage of the shoot, adventitious roots send forth. The shoot of astrain of high-quality Chinese mugwort is collected, and undergoes cutting propagation so as to grow into a seedling; the genotype of the seedling are identical to that of the stock plant, and a clone with excellent cultivated characters can be planted; and yield increase and high quality characteristics can be given to full play; and genetic differentiation can be avoided. With the Chinese mugwort cutting propagation method of the invention adopted, a large number of plants with the same genotype can be propagated through cottage, namely a stock plant can be cloned, and economic benefits canbe maximized.
Owner:BEIHUA UNIV
Agrobacterium tumefaciens-mediated sugarcane callus efficient genetic transformation method
ActiveCN114774464AImprove transformation activityImprove accuracyFermentationPlant tissue cultureBiotechnologyAntioxidant
The invention discloses an efficient genetic transformation method for agrobacterium-mediated sugarcane calluses, and belongs to the field of tissue culture. According to the invention, by enhancing the Agrobacterium tumefaciens Vir gene induction activation level and the bacterial liquid dip dyeing strength of sugarcane calluses, the occurrence frequency of transgenic events is greatly improved; an antioxidant is added into subsequent co-culture, screening and differential culture media to inhibit callus browning caused by agrobacterium-enhanced dip dyeing, so that the survival rate and the differentiation capacity of the dip-dyed calluses are obviously improved; a double-marker screening system with a green fluorescent protein marker EGFP gene and a herbicide glufosinate-ammonium resistance marker Bar gene at the same time is adopted, positive transgenic calluses are visually and nondestructively screened by using an EGFP marker, positive transgenic differentiated seedlings are screened by using Bar marker resistance, the accuracy of transgenic event screening is remarkably improved, and the screening efficiency is improved. And finally, the efficient genetic transformation of the agrobacterium tumefaciens mediated sugarcane callus is realized.
Owner:SUGARCANE RES INST OF YUNNAN ACADEMY OF AGRI SCI
Quadruple fluorescent SSR molecular marker detection method for tea tree
InactiveCN108998551AAvoid contactIncrease the number of testsMicrobiological testing/measurementResearch efficiencyFluorescence
The invention discloses a quadruple fluorescent SSR molecular marker detection method for a tea tree. According to the invention, a set of detection method for quadruple SSR molecular markers in the tea tree is established by the combination of a primer combination, a touchdown PCR (polymerase chain reaction), capillary fluorescence electrophoresis and other technologies, and the method can not only improve the research efficiency and accuracy of the SSR molecule markers in the tea tree, but also save labor. However, the current detection of the SSR molecular markers in the tea tree mainly adopts a polyacrylamide gel electrophoresis technology, which has the disadvantages of low resolution, large error, low efficiency and the like; and by adopting the quadruple SSR molecular marker detection method provided by the invention, the detection of 4 pairs of SSR primers can be completed at a time. At the same time, the defects of primer annealing temperature screening, manual running rubber,artificial reading tape and the like are avoided, not only is the work efficiency greatly improved, but also the method is simple, rapid, accurate, stable and good in repeatability, and can be used for the research of tea tree variety identification, fingerprint mapping construction, population genetic structure, genetic differentiation and the like.
Owner:湖南省茶叶研究所
SSR fluorescent-labeled primers for paternity identification and population genetic analysis of the Great Salamander
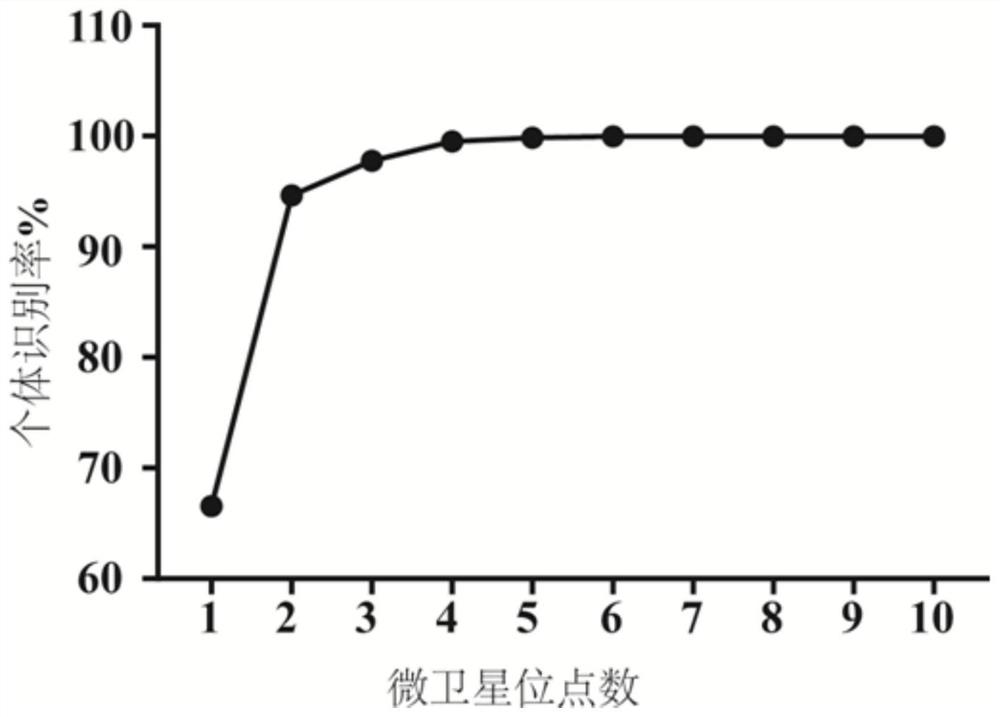
ActiveCN112501315BEasy to readStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationStructure analysisMicrosatellite
The invention belongs to the field of genetics, and in particular relates to SSR fluorescent-labeled primers for paternity identification and population genetic analysis of the great liang salamander. The specific technical plan is: a set of methods for individual identification, paternity identification, and population genetic structure analysis of the salamander and 10 pairs of microsatellite primers. The sites (primers) provided by the invention have both accuracy and polymorphism. By applying the technology provided by the invention to population selection research and population genetic analysis, scientifically feasible protection suggestions can be put forward from the perspectives of reproduction and genetic differentiation, laying the foundation for future conservation work.
Owner:CHENGDU INST OF BIOLOGY CHINESE ACAD OF S
Camphor tree whole genome ssr molecular marker and its preparation method and application
ActiveCN108823327BAvoid missingMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyGene flow
The invention discloses a whole genome SSR molecular marker of camphor tree and its preparation method and application. The camphor tree microsatellite molecular markers developed by the present invention can be applied to the population genetic diversity detection and kinship analysis of ancient camphor trees, providing a new batch of microsatellite molecular markers for Lauraceae plants, and also for the population genetic differentiation of Lauraceae plants New tools are provided for the study of genetic diversity and structure, the level of genetic diversity of populations, the gene flow and mating system between populations, species evolution, and molecular assisted breeding.
Owner:JIANGXI ACAD OF FORESTRY
Method for determining genetic differentiation between rice mixed interplanting varieties by SSR markers
ActiveCN111719011ASolve complexitySolve the problem of selectivityMicrobiological testing/measurementClimate change adaptationBiotechnologyElectrophoreses
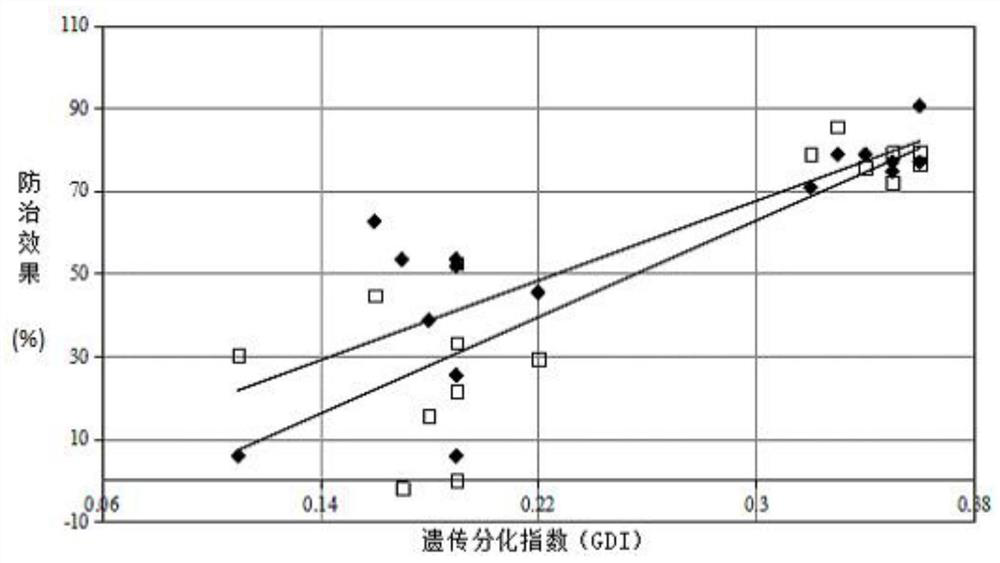
The invention relates to a method for determining genetic differentiation between rice mixed interplanting varieties by SSR markers, and belongs to the technical field of molecular markers. The methodcomprises the following steps: extracting total DNA from to-be-detected rice, performing amplifying by using 48 pairs of SSR specific primers, separating a PCR product by using a capillary electrophoresis method, reading the molecular weight of the PCR product, and carrying out datamation treatment on SSR molecular fingerprints, and calculating a genetic differentiation index (GDI) according to agenetic differentiation formula; and along with the increase of the genetic differentiation index GDI, in the diversified mixed interplanting planting, the overall rice blast control effect of the variety matching combination being better. According to the method, the genetic differentiation level among varieties can be rapidly and accurately identified by using a trace amount of DNA samples, thecontrol effect of diversity mixed interplanting assembled varieties on rice blast is predicted, and the method has important significance in research of variety matching selection in diversity mixedinterplanting.
Owner:YUNNAN AGRICULTURAL UNIVERSITY
SSR fluorescence labeling primers for paternity test and population genetic analysis of Liangshantriton taliangensis
ActiveCN112501315AEasy to readStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationGeneticsBioinformatics
The invention belongs to the field of genetics, and particularly relates to SSR fluorescence labeling primers for paternity test and population genetic analysis of Liangshantriton taliangensis. According to the specific technical scheme, a group of methods capable of performing individual identification, paternity test and population genetic structure analysis of the Liangshantriton taliangensis and 10 pairs of microsatellite primers are provided. The loci (primers) provided by the invention have the accuracy and polymorphism. When the technology provided by the invention is used for population selection research and population genetic analysis, scientific and feasible protection suggestions can be provided from the perspectives of propagation, genetic differentiation and the like, and a foundation is laid for future conservation work.
Owner:CHENGDU INST OF BIOLOGY CHINESE ACAD OF S
Screening method and application of Henan gamecock genome SNP molecular markers
ActiveCN109207610BComprehensive and effective germplasm analysisImprove detection accuracyMicrobiological testing/measurementGermplasmGenetic divergence
The invention relates to the field of genetics, in particular, it provides a screening method of Henan gamecock genome SNP molecular marker and its application. The screening method detects through high-throughput sequencing, calculates genetic statistics, analyzes the genetic differences between Henan gamecock and other chicken breeds, and screens out Henan gamecock SNP molecular markers. This screening method alleviates the technical problems of high cost and low accuracy of genetic differentiation research caused by the lack of efficient molecular biology methods and genetic markers in the identification of genetic differentiation of gamecocks in Henan, and achieves the technical effect of efficient and rapid screening of molecular markers in Henan gamecocks , the selected genes of Henan Gamecock were screened and used as molecular markers to identify the germplasm of Henan Gamecock, with low cost, abundant data and high detection accuracy. The application of the SNP molecular markers for the identification of Henan gamecock genome has alleviated the technical problem of lack of efficient molecular biology markers in the study of genetic differentiation of Henan gamecock.
Owner:JIANGSU INST OF POULTRY SCI
Method for domesticating desert plant agriophyllum squarrosum from beginning based on population polymorphism
InactiveCN112205247AImprove adaptabilitySpeed up gene flowSeed and root treatmentCultivating equipmentsSoil typeGene flow
The invention relates to the technical field of domesticating desert plants from the beginning, in particular to a method for domesticating desert plant agriophyllum squarrosum from the beginning based on population polymorphism. The method comprises the steps that phenotypic variation caused by soil type, slope, climate and hydrological environment difference is excluded through homogeneous garden experiments; morphological or phenotypic differences formed by genetic differentiation is screened out through phenotypic data determination of wild agriophyllum squarrosum; the optimal natural growth condition influencing the heritable phenotype of the agriophyllum squarrosum is determined by combining the meteorological factor data of the agriophyllum squarrosum group of the origin; and groupgenetic evaluation is carried out on domesticated high-quality germplasm resources, and large-area popularization and demonstration of the germplasm resources are determined. According to the method for domesticating the desert plant agriophyllum squarrosum from the beginning based on population polymorphism, a genetic database of germplasm resource populations is constructed for the first time, gene flow among the populations is accelerated through the homogeneous garden experiments, and the adaptability of crop target phenotypes to the climate is improved; and the traditional time-consumingand labor-consuming artificial hybridization process is changed, and various risks caused by a transgenic technology are further avoided.
Owner:NANTONG UNIVERSITY
The ssr primer pair of persimmon anthracnose based on the genome of related species and its application
ActiveCN106701943BEffective prevention and controlMicrobiological testing/measurementMicroorganism based processesBiotechnologyMicrobiology
The invention belongs to the field of biotechnology, and specifically discloses a set of SSR primer pairs for persimmon anthracnose fungus developed based on the genome of a related species and an application thereof. The SSR primer pair of the persimmon anthracnose fungus is based on the genome of a close relative of the persimmon anthracnose fungus—the fruit-bearing anthracnose fungus, and 30 pairs of SSR primers have been developed through MISA software. The screening of pathogens has obtained 15 pairs of SSR primers with polymorphisms. The primers for Kaki anthracnose bacteria developed by the present invention are new markers that exist stably and can be used for the analysis of genetic diversity and genetic differentiation of Kaki anthracnose bacteria. It can also be used in the identification of persimmon anthracnose species and the study of kinship, which is of great significance to the effective prevention and control of persimmon anthracnose.
Owner:河南省林业科学研究院
SSR (Simple Sequence Repeat) molecular marker for genetic diversity analysis of clematis nepalensis and application of SSR molecular marker
The invention discloses an SSR (Simple Sequence Repeat) molecular marker for genetic diversity analysis of clematis nepalensis and application of the SSR molecular marker. The invention also discloses an SSR primer group for genetic diversity analysis of clematis malaytea, and the SSR primer group consists of a primer pair 1-a primer pair 29. The SSR primer group has any one of the following functions of n1)-n5): n1) analyzing or evaluating genetic diversity of clematis malaytea; n2) analyzing or evaluating the genetic differentiation degree of the clematis nepalensis; n3) analyzing or evaluating the genetic relationship of the clematis malaytea; n4) constructing or preparing a heredity map or fingerprint spectrum of the clematis nepalensis; and n5) identifying or distinguishing different geographical populations of clematis maple. Sequencing results of simplified genomes are used for splicing, the SSR molecular marker suitable for research on the genetic diversity of the clematis nepalensis is screened out, and the SSR molecular marker has important significance on development of the research on the genetic diversity of the clematis nepalensis and conservation work.
Owner:北京市园林绿化科学研究院
Duolang sheep SNP marker and its screening method and application
ActiveCN106011259BImprove accuracyOvercoming the pitfalls of phenotypingMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyGenetic linkage disequilibrium
The invention discloses a Duolang sheep SNP (single-nucleotide polymorphism) marker, and a screening method and application thereof, belonging to the field of animal molecular markers. A genetic differentiation coefficient method is utilized to perform screening on a commercialized sheep genome SNP chip and further remove high-linkage-disequilibrium SNP markers (r<2>>0.2) through linkage disequilibrium analysis, thereby obtaining the Duolang sheep SNP marker. The Duolang sheep SNP marker can be applied to identification of Duolang sheep species or animal products thereof on the premise of no obvious linkage disequilibrium among markers; and therefore, when the Duolang sheep SNP marker disclosed by the invention is used for identifying the Duolang sheep species or animal products thereof, no interference among sites exists, thereby greatly enhancing the Duolang sheep species identification accuracy and effectively lowering the identification cost.
Owner:新疆畜牧科学院生物技术研究所
Polymorphic primers of cinnamomum camphora chloroplast SNP molecular markers and application of polymorphic primers
ActiveCN110760610AFully reveal the true levelGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationCinnamomum camphoraGene flow
The invention discloses polymorphic primers of cinnamomum camphora chloroplast SNP molecular markers and application of the polymorphic primers, and belongs to the technical field of forestry molecular biology. 11 pairs of polymorphic primers of the cinnamomum camphora chloroplast SNP molecular markers are screened and applied to ancient cinnamomum camphora genetic diversity analysis and ancient cinnamomum camphora pedigree analysis, and on the basis of chloroplast haplotype analysis, results are shown that different haplotypes have obvious distribution differences, most haplotypes only existin specific populations, and only few haplotypes are shared haplotypes; and thus a fact is indicated that certain geographic differentiation exists among the haplotypes. A genetic differentiation coefficient Fst of 18 cinnamomum camphora populations is 0.212, gene flow Nm of the cinnamomum camphora populations is 0.929, and a result is shown that middle gene flow exists among the cinnamomum camphora populations. By adopting the polymorphic primers, the SNP molecular markers have good repeatability, a true level of cinnamomum camphora genetic diversity can be revealed completely, and theoretical foundations are provided for wild cinnamomum camphora resource protection and utilization.
Owner:NANJING FORESTRY UNIV +1
A method for predicting the seed-setting rate of indica-japonica rice hybrid f1 using indel molecular markers
InactiveCN103798127BGuaranteed outputMicrobiological testing/measurementPlant genotype modificationBiotechnologyJaponica rice
The invention belongs to the technical field of molecular mark assisted breeding, and particularly discloses a method for detecting genetic differentiation level of indica-japonica hybridizing parents and predicting the percentage of fructification of hybrid F1 by using InDel molecular markers. The hybridizing parents can be purposefully selected and the hybrid F1 with high percentage of fructification is prepared by detecting the genetic differentiation level of the indica-japonica hybridizing parents. The indica-japonica genetic differentiation coefficients (GDI) of indica-japonica hybridizing parents are determined by using 40 InDel molecular markers and a built calculation method, the correlation between the indica-japonica hybridizing parents GDI and a really measured mean value of the percentage of fructification (PF) of the hybrid F1 is really analyzed, so as to form a method for analyzing parents GDI to predict the percentage of fructification of the indica-japonica hybrid F1 by the molecular markers. The indica-japonica genetic differentiation coefficient detection on the parents can be carried out before hybridization, so as to predict the percentage of fructification of the hybrid F1. Thus, the indica-japonica hybridizing parents can be effectively selected.
Owner:FUDAN UNIV
Microfluidic system and method of use thereof
ActiveUS20190316075A1Cost efficientEffective timeBioreactor/fermenter combinationsValve arrangementsHospital basedCell type specific
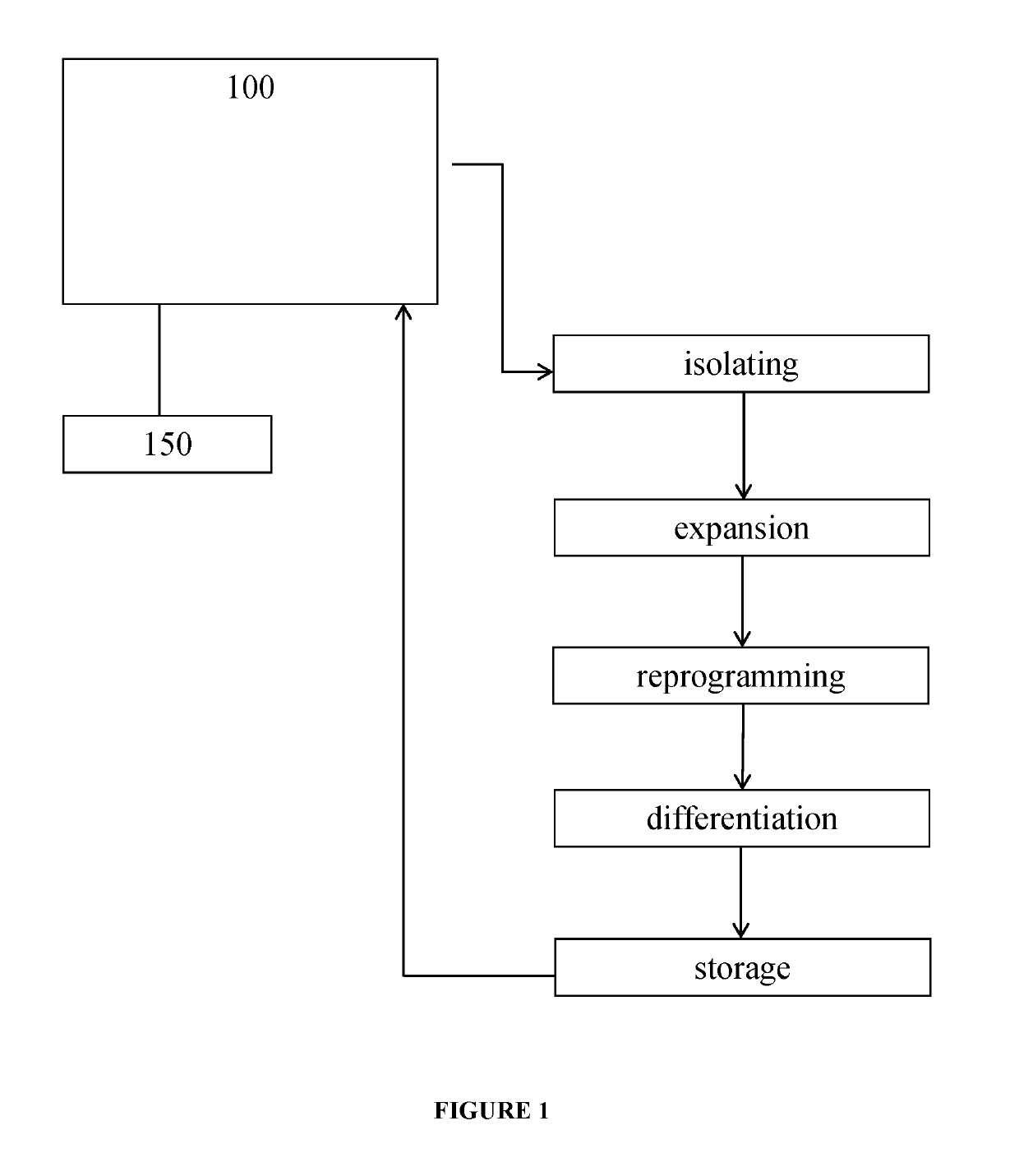
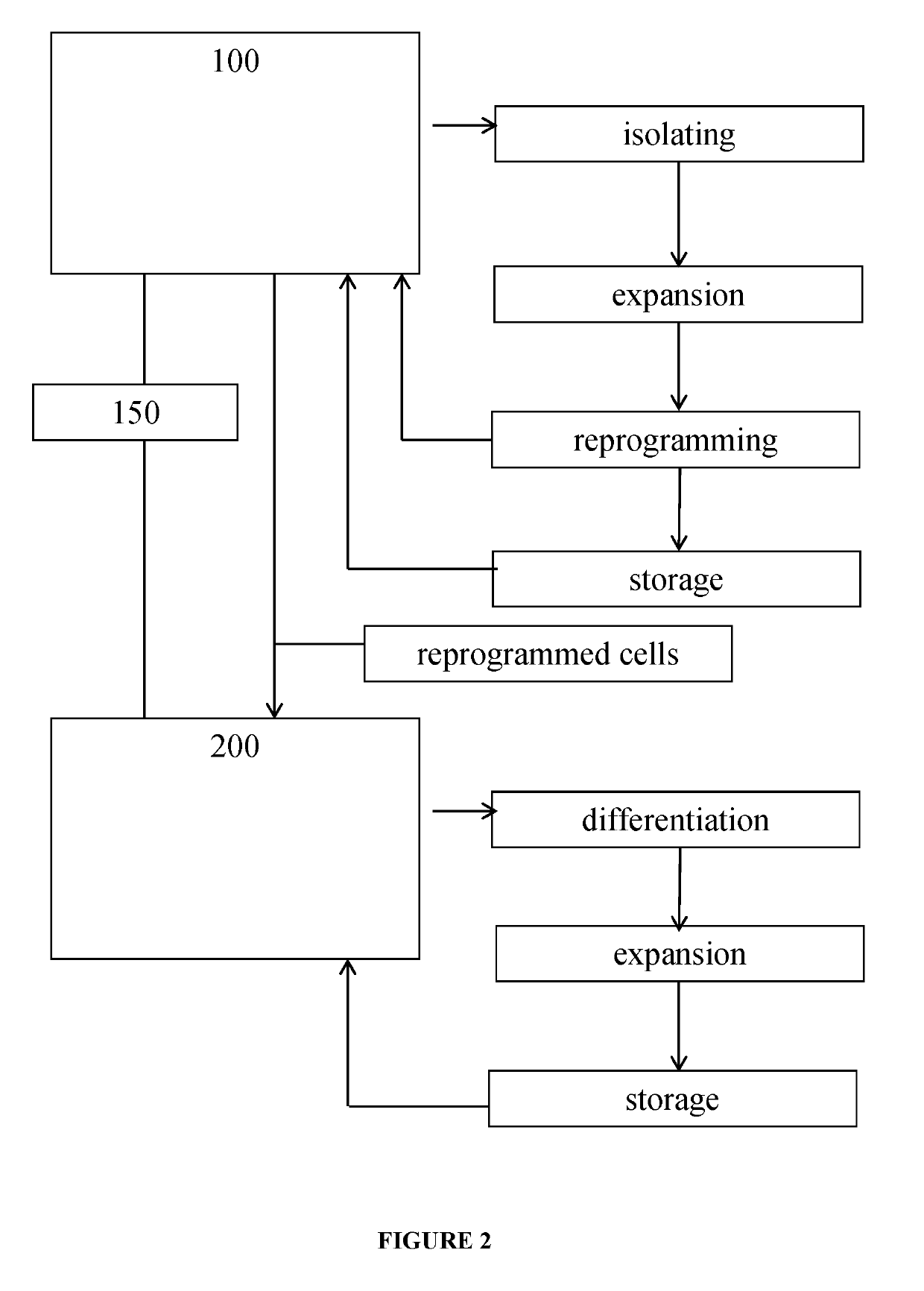
This invention concerns an integrated microfluidic system that utilizes microfluidic chip technology to receive a patient sample including cells, expand the cells, reprogram the expanded cells and then store the reprogrammed cells in a microfluidic chip. These microfluidic chips with stored reprogrammed cells may then be used in scenarios of genetic differentiation into specific cell types. Overall this system and workflow is suitable as a hospital based device that will allow the generation of iPSCs from every isolating patient for downstream diagnostic or therapeutic use.
Owner:NEW YORK STEM CELL FOUND INC
SSR primers of endangered Heritiera littoralis Hernandia nymphaeifolia and application thereof
ActiveCN114015802AIncrease diversityMicrobiological testing/measurementBiostatisticsMangrove plantsGenetic architecture
The invention belongs to the field of molecular genetics, and discloses application in the aspects of genetic diversity, genetic structure analysis and the like of Hainan Island Hernandia nymphaeifolia populations developed on the basis of SSR molecules, wherein genetic structure characteristics and genetic diversity of different populations of Hernandia nymphaeifolia are analyzed, gene exchange and genetic differentiation degrees among different populations are clarified, phylogenetic evolution and geographical differentiation of endangered Heritiera littoralis Hernandia nymphaeifolia populations are clarified, a foundation is laid for molecular biology and genetics research of the endangered Heritiera littoralis Hernandia nymphaeifolia, and a theoretical basis is provided for protection of genetic diversity of Hernandia nymphaeifolia, effective utilization of resources and the like. According to the invention, the SSR marker developed by adopting the transcriptome high-throughput technology is high in speed, high in efficiency and low in cost.
Owner:HAINAN NORMAL UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com