SSR (Simple Sequence Repeat) molecular marker for genetic diversity analysis of clematis nepalensis and application of SSR molecular marker
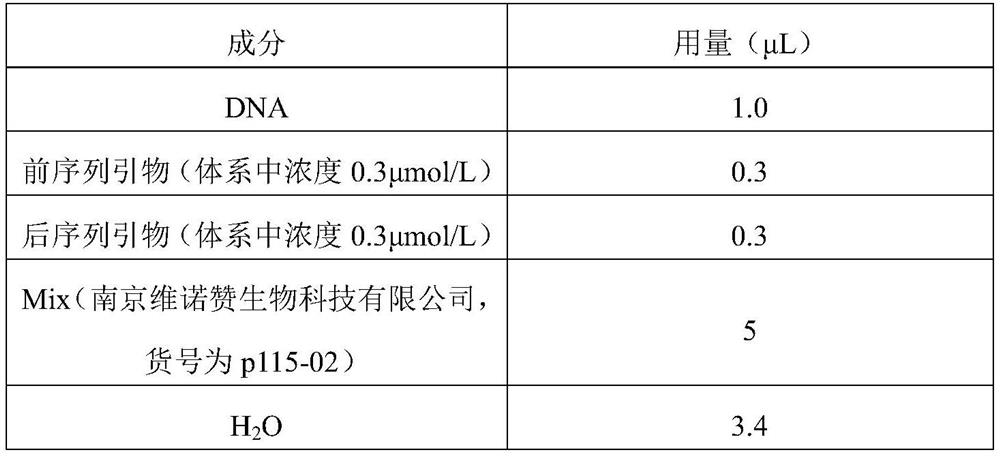
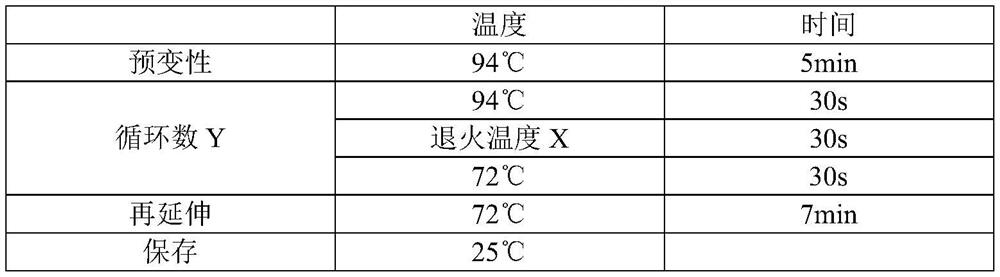
A DNA molecule, 2-R technology, applied in the direction of sequence analysis, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0152] Example 1, Screening of Clematis maple leaf SSR molecular markers
[0153] 1. Test materials and methods
[0154] 1. Test material
[0155] Clematis maple leaf is mainly distributed in Beijing area, and occasionally distributed in Hebei and Henan. According to the field investigation results, the following nine locations were selected for sampling: Sibeiyu (Hebei), Gougezhuang (Hebei), Fangshan Sandu (Beijing), Wuheer Tunnel (Beijing), Yuntai Mountain (Henan), Nanshi Yangyang Grand Canyon (Beijing), Jingxi Ancient Road (Beijing), Xiayunling (Beijing), and Xinghuang (Beijing), the sampling standard is to sample 10 plants for each population, and the distance between individual plants should be at least 2m. Avoid taking samples from the same cliff crevice s material. The relevant information and numbers of all test materials are shown in Table 1.
[0156] Table 1. Basic information of population sampling
[0157] group number Place Number of samples S...
Embodiment 2
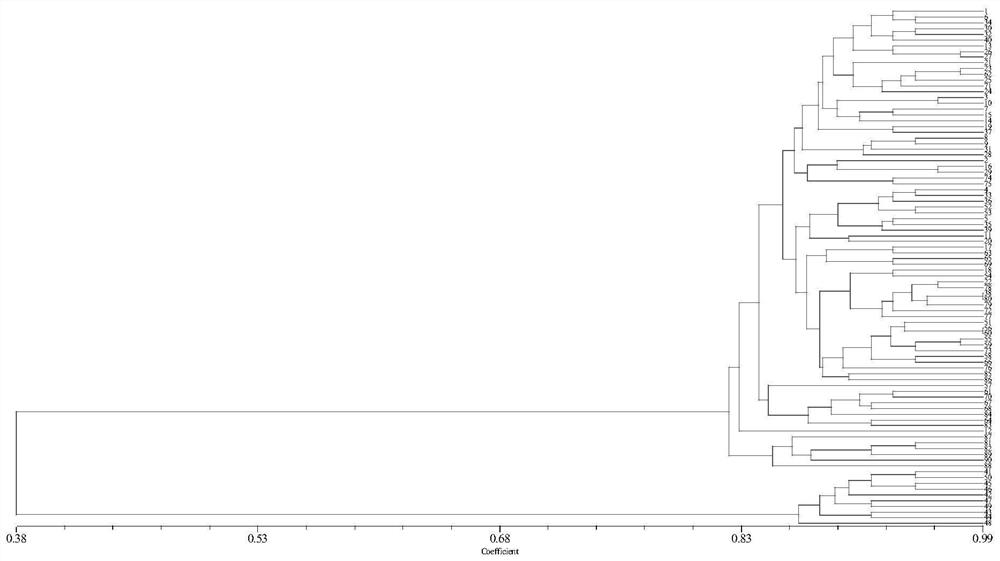
[0181] Example 2. Using SSR molecular markers to analyze the genetic diversity of Clematis maple leaf
[0182] 1. Genetic diversity parameters of 29 SSR loci
[0183] Observed allele number (Na), effective allele number (Ne), Shannon's diversity index (I), observed heterozygosity (Ho), and expectation of 29 SSR loci using population genetics software POPGENE Version1.32 Heterozygosity (He) and F statistics (Fit, Fis, Fst) were calculated. Polymorphism information content (PIC) was calculated using PowerMarker V3.25 software. Gene flow (Num)=(1-Fst) / 4Fst was calculated.
[0184] The results are shown in Table 5. It can be seen from Table 5 that in the 9 populations, the observed allele numbers (Na) of the 29 SSR loci varied from 2 to 4, with an average value of 2.4483. The effective allele number (Ne) ranged from 1.1172 to 2.2839, with an average of 1.6310. Shannon's diversity index (I) ranged from 0.2146 to 1.0092, with an average value of 0.5682; the primer with the highe...
Embodiment 3
[0193] Example 3. Using SSR molecular markers to calculate the genetic differentiation coefficient Fst among 9 Clematis maple leaf populations
[0194] The genetic differentiation factor (Fst) is an indicator of the degree of differentiation between populations and varies between 0 and 1. Fst less than 0.05 indicates that there is basically no genetic differentiation trend between the two populations, Fst between 0.05 and 0.15 indicates a moderate degree of differentiation, Fst between 0.15 and 0.25 indicates a large degree of genetic differentiation, and Fst greater than 0.25 indicates the degree of differentiation very big. The genetic differentiation coefficients of nine Clematis maple populations were calculated using GenALEx 6.5.
[0195] The results are shown in Table 7. It can be seen from Table 7 that the genetic differentiation coefficient (Fst) among the nine Clematis maple leaf populations varies from 0.023 to 0.469. The genetic differentiation coefficient (Fst) o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com