Patents
Literature
289 results about "Population genetics" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Population genetics is a subfield of genetics that deals with genetic differences within and between populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as adaptation, speciation, and population structure.
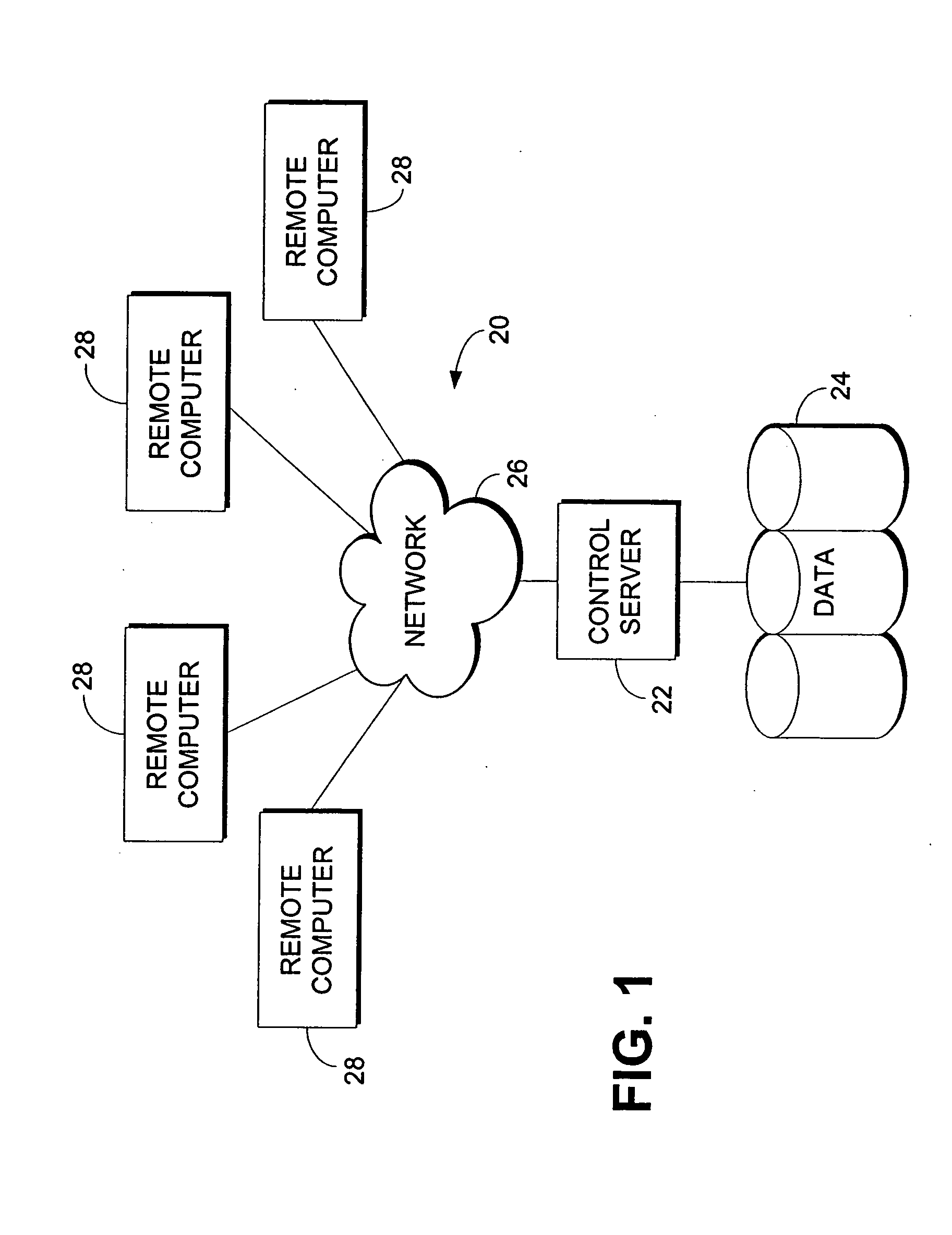
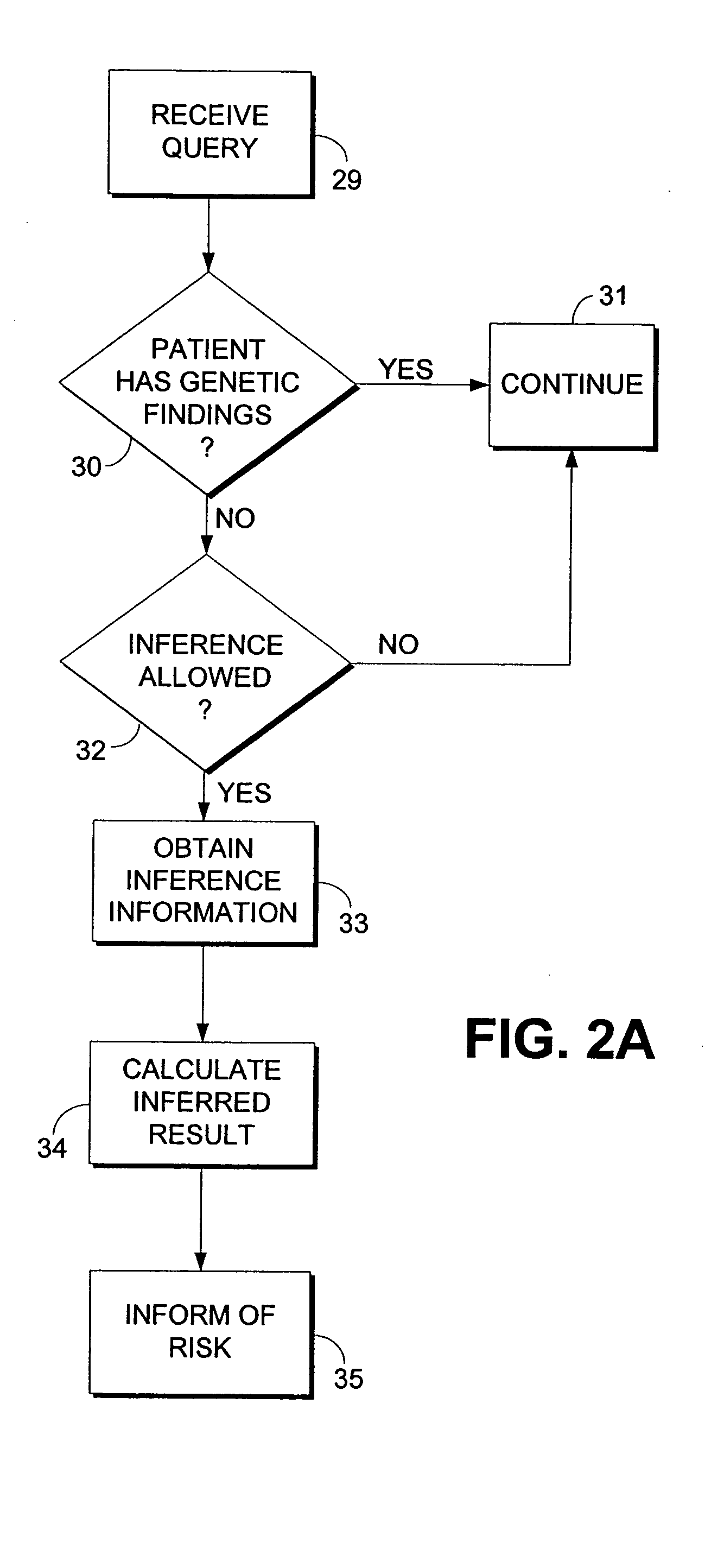
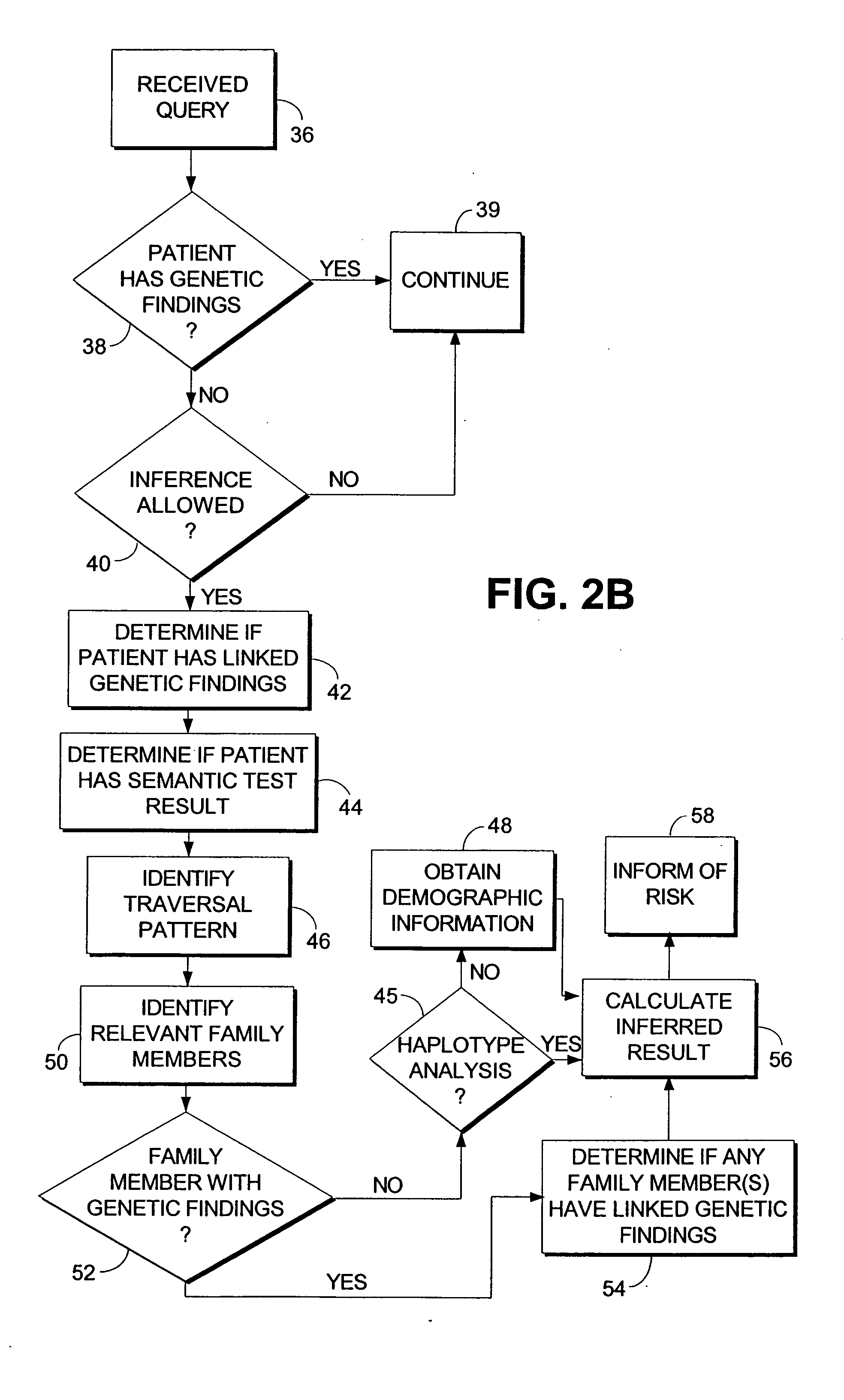
Computerized method and system for inferring genetic findings for a patient
A method and system in a computing environment for inferring genetic findings for a patient is provided. The method includes receiving a request for genetic findings for a person from another application or a user. The method further includes inquiring as to whether the person has the genetic findings. If not, the method automatically provides inferred genetic findings for the person. The inferred genetic findings are calculated using genetic findings for family members of the patient, linkage analysis, haplotype analysis, semantic test results for the person and / or population genetics information.
Owner:CERNER INNOVATION
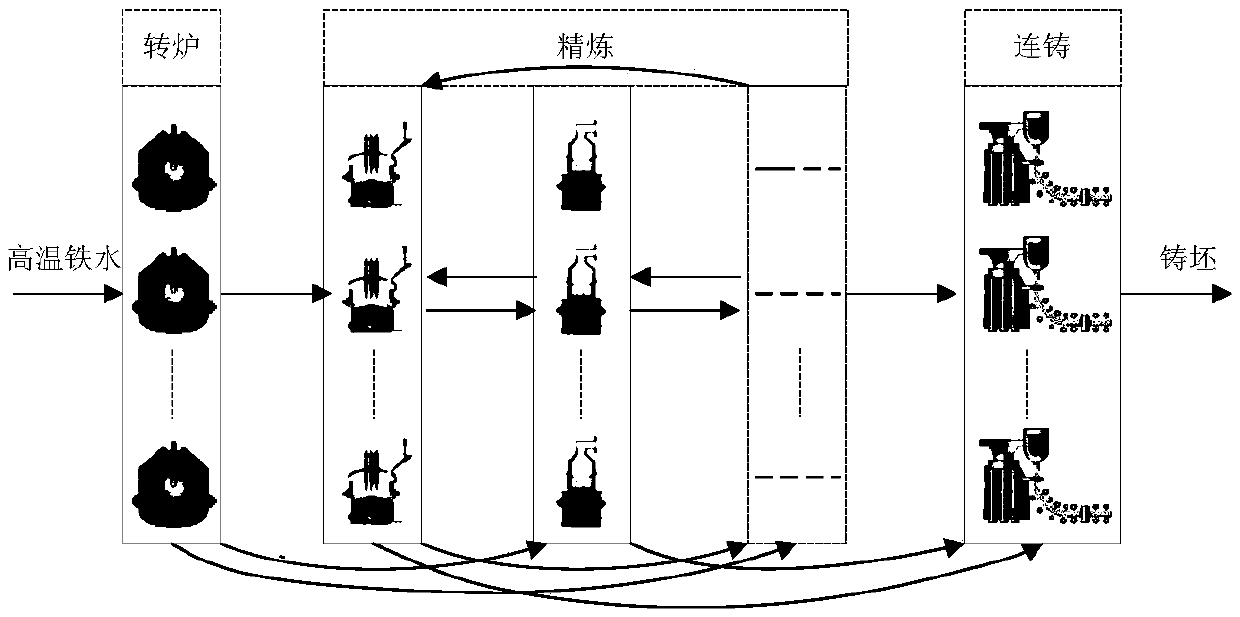
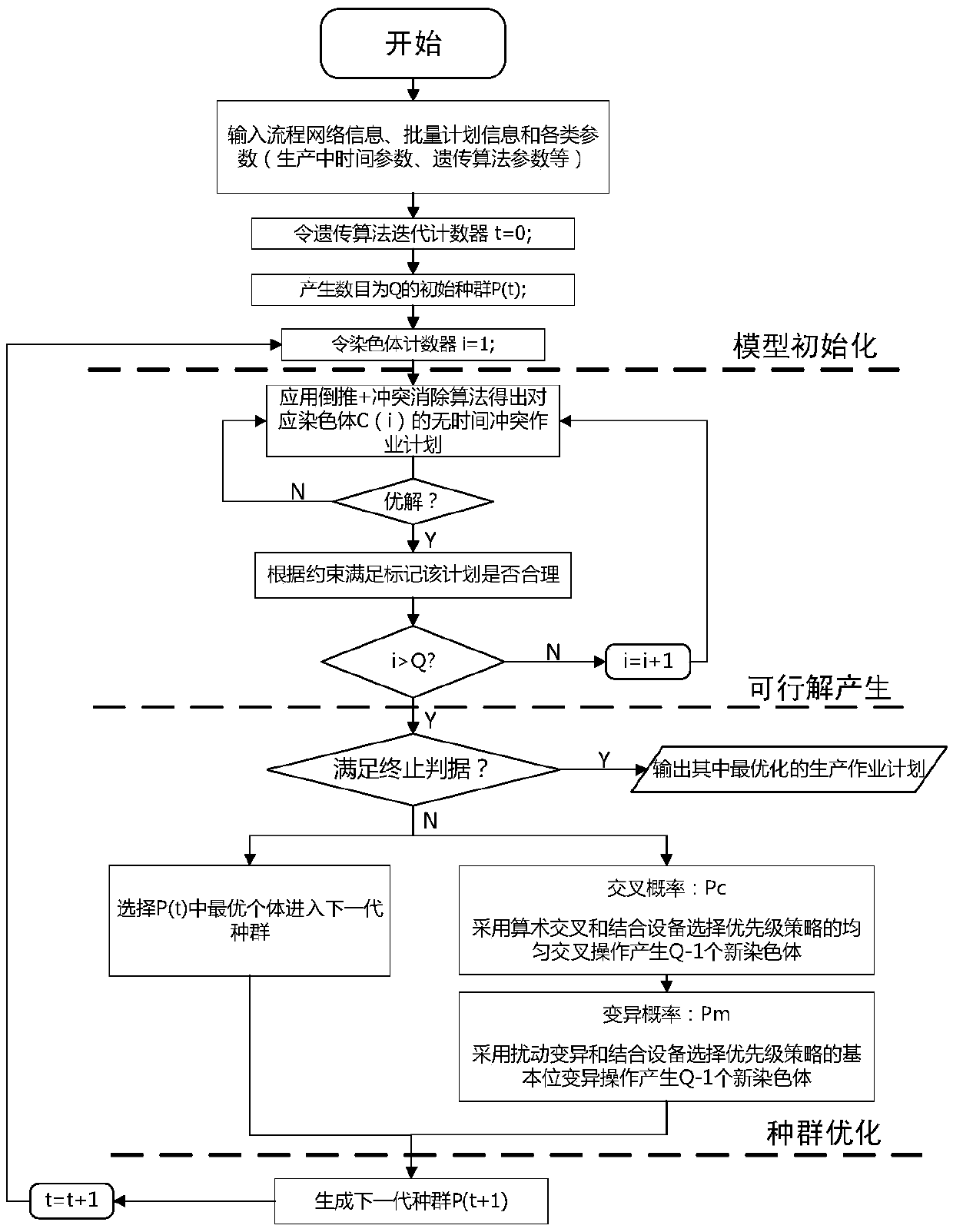
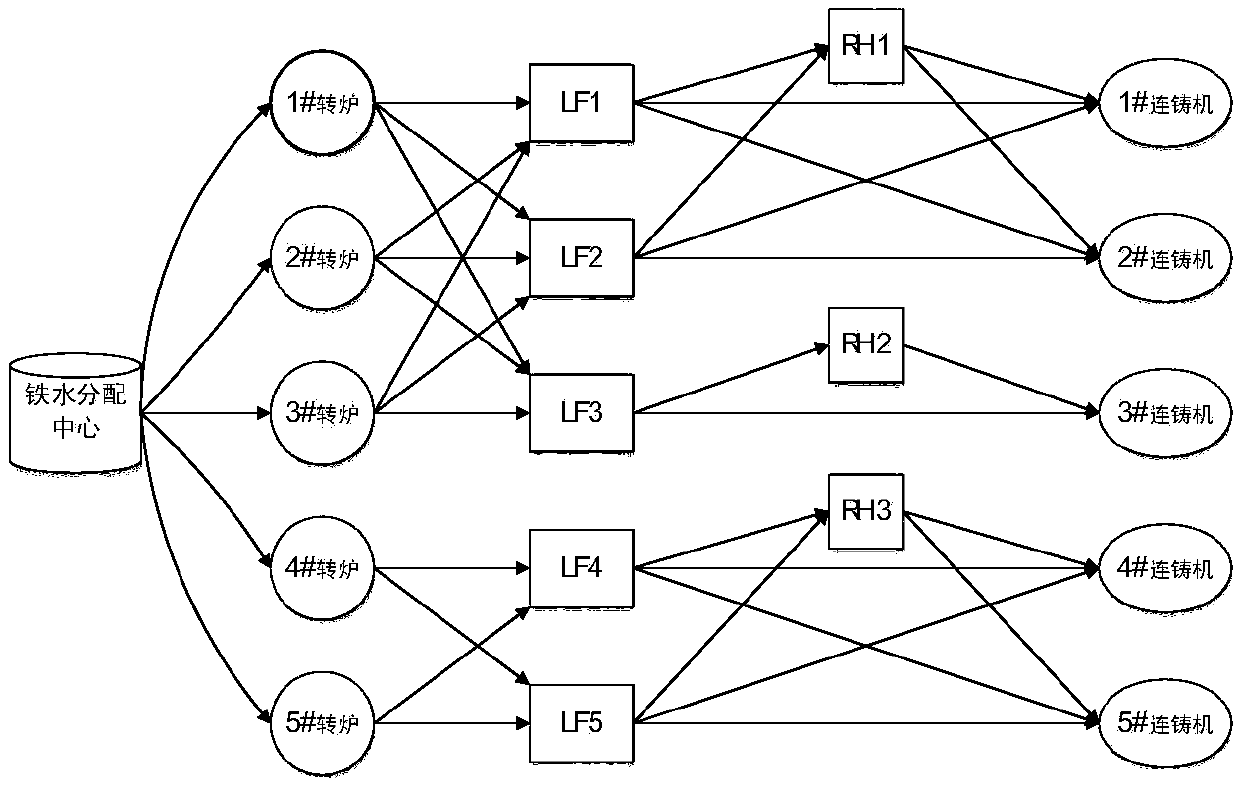
Steelmaking-continuous casting scheduling method utilizing priority policy hybrid genetic algorithm
The invention proposes a steelmaking-continuous casting scheduling method utilizing a priority policy hybrid genetic algorithm. The method comprises the following steps: establishing a production scheduling plan target function; establishing a constraint condition set; performing iterative calculation on the target function by utilizing the priority policy hybrid genetic algorithm; and calculating decision variables. The calculation of the decision variables specifically comprises the steps of performing model initialization; calculating a feasible solution: designing a segmented combined real number code composed of casting time information of a continuous casting machine and information of a heat machining device, randomly generating operation time according to a distribution law, and obtaining a time conflict-free scheduling plan by backward inference calculation and conflict elimination methods; and performing population genetic optimization: quantitatively describing a matching relationship among machining devices in reality by using processing weight assignment of a task executable device, and introducing the matching relationship to genetic operation in the form of a device selection priority policy to perform population evolution. The method can solve the problem of uncertainty of device selection and operation time in production to obtain an optimized executable production scheduling plan.
Owner:CHONGQING UNIV
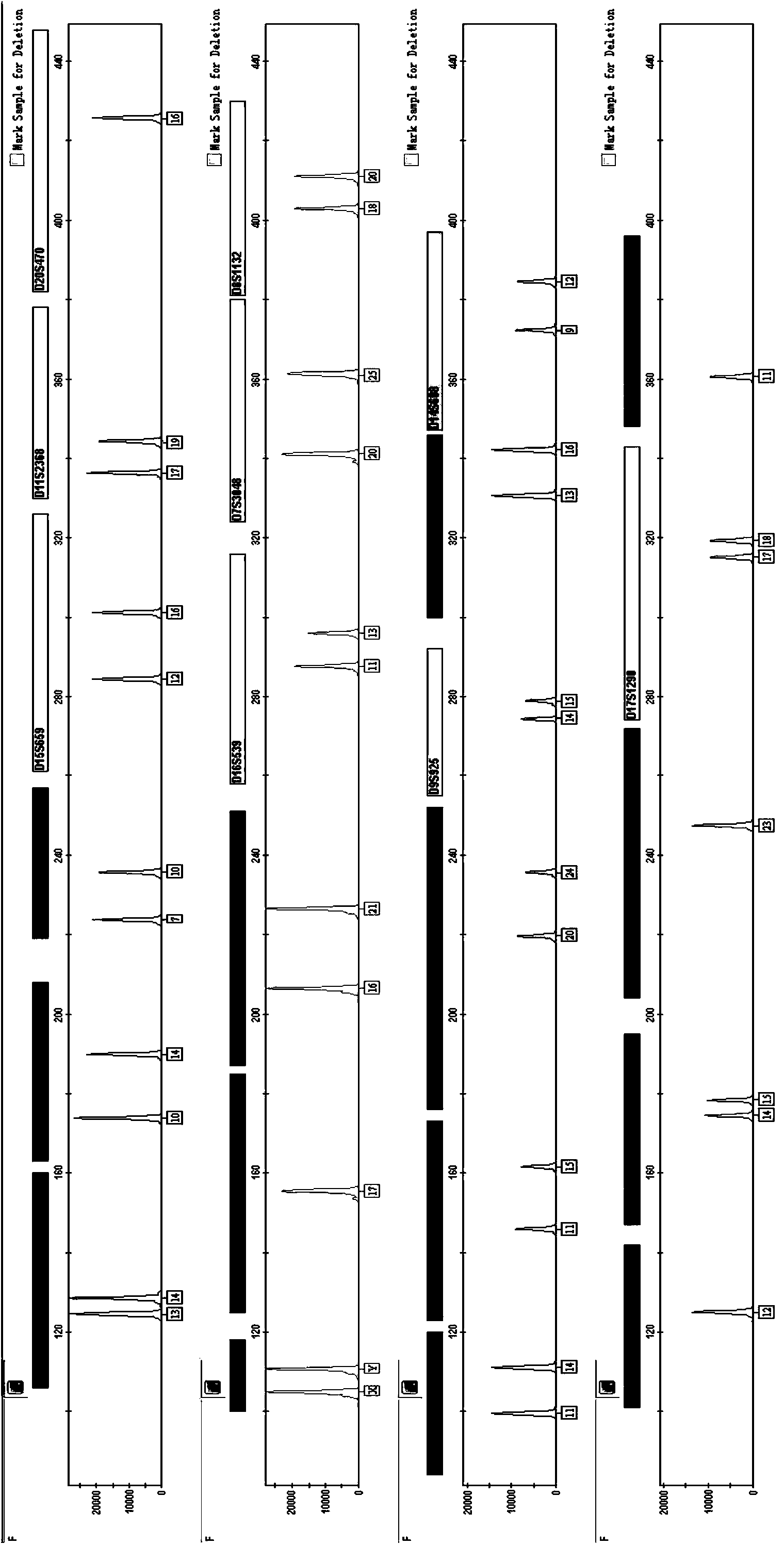
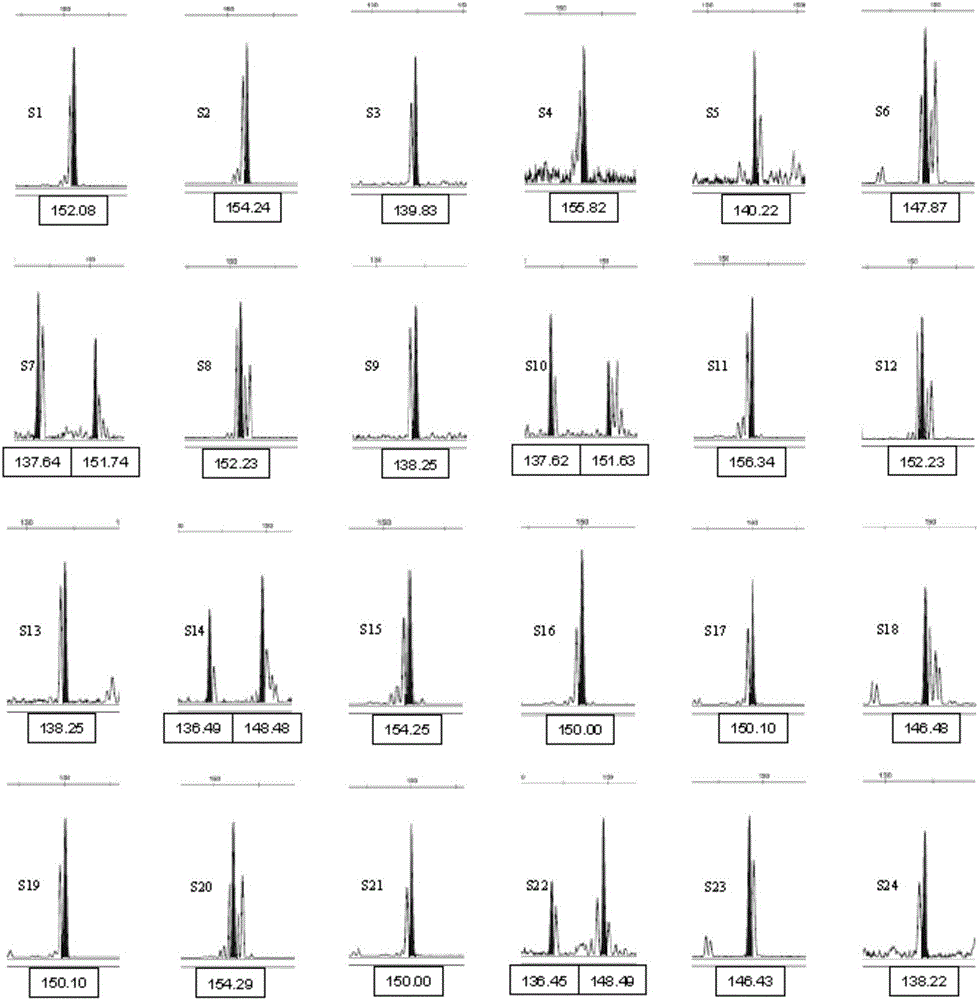
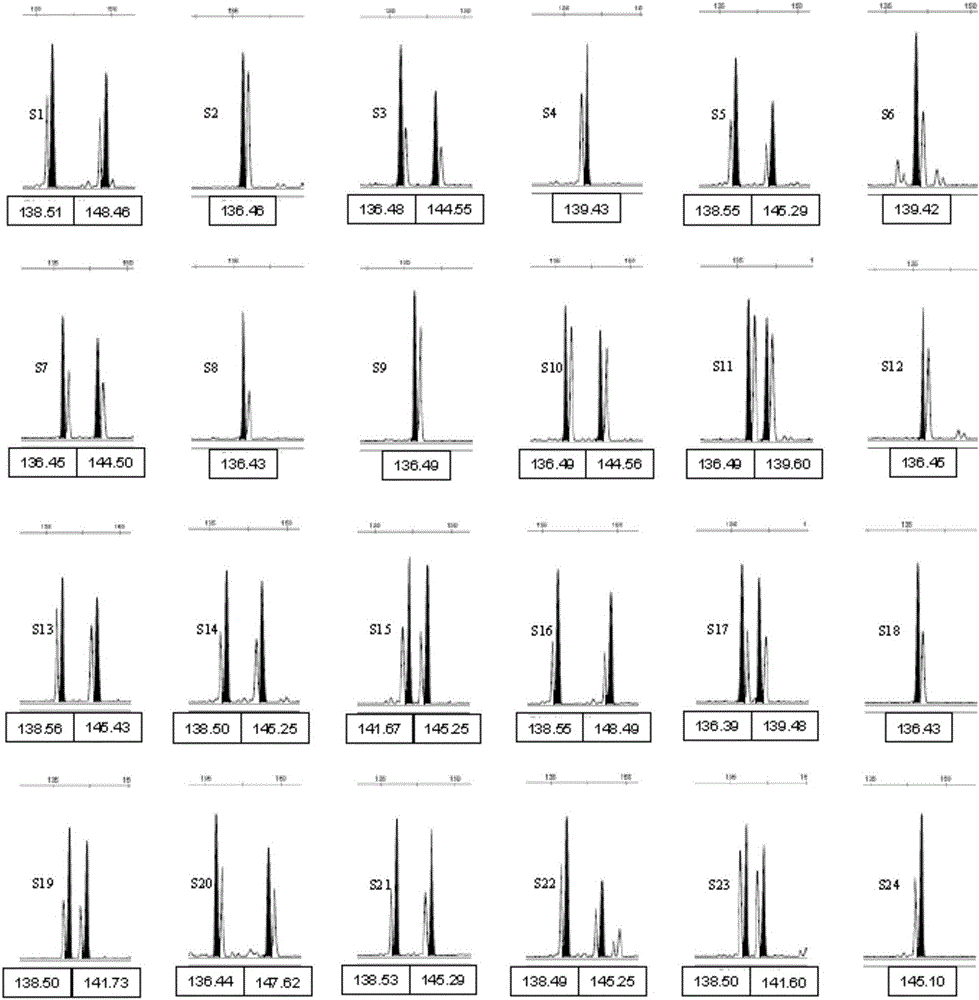
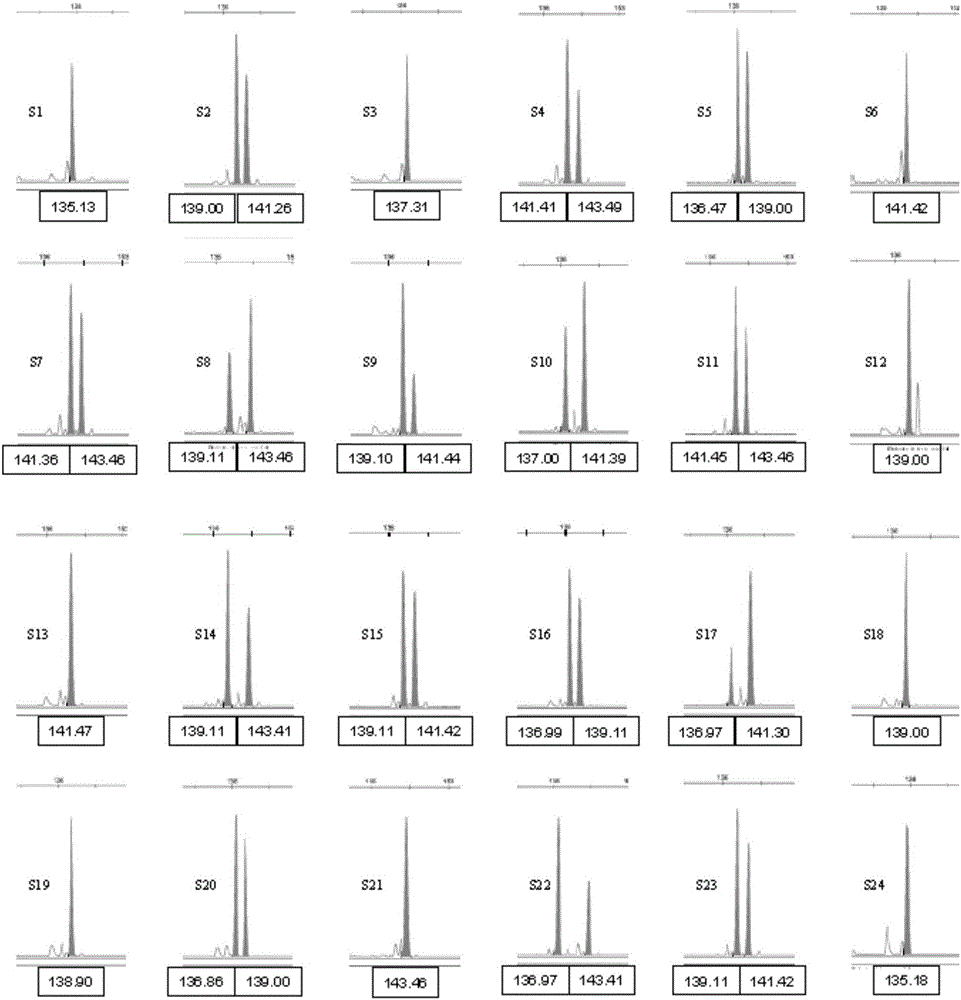
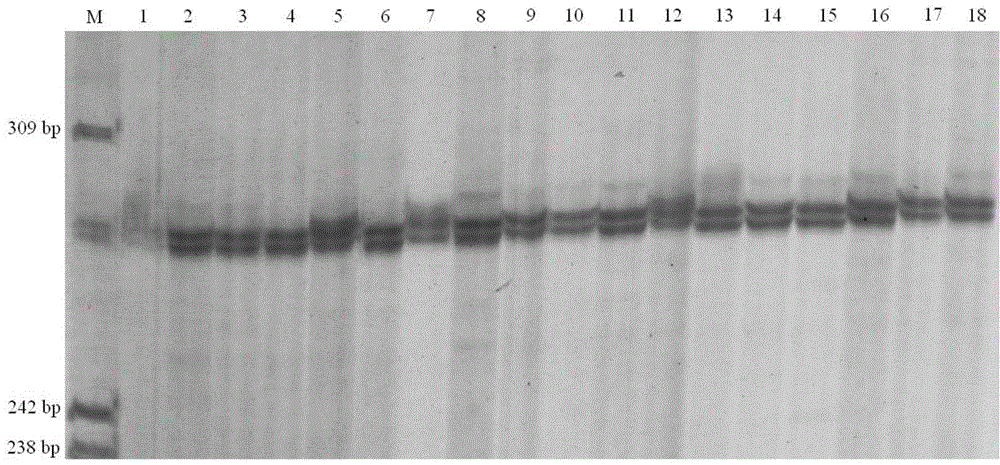
Composite amplification system of 23 short tandem repeat sequences and a kit
ActiveCN103834732AImprove performanceIncrease independenceMicrobiological testing/measurementBiotechnologyDNA paternity testing
The invention relates to a composite amplification system of 23 short tandem repeat sequences and a kit, is used for detecting heredity mark gene of polymorphism in a mankind genome, and belongs to the biology technical field. The invention relates to a scheme that a plurality of short tandem repeat sequences are simultaneously amplified in a PCR system; a specific gene locus comprises 22 short tandem repeat sequences with high level heredity polymorphism and 1 gender determination locus; primers are respectively designed and a fluorophore is marked; the system has very high individual recognition rate and non-father elimination rate; the system and kit can be used for legal medical individual recognition, paternity tests, and colony genetics analysis, is high in accuracy, and good in sensitivity.
Owner:SUZHOU MICROREAD GENETICS
Method of screening pacific oyster EST micro-satellite mark
The invention discloses a screening method of Pacific oyster EST micro satellite label, which comprises the following steps: finding micro satellite site with Pacific oyster EST sequence announced by GeneBank and micro satellite searching software SSRhunter; screening and separating 2-6 base repeat unit micro satellite fragment (repeat times >5); getting repeat ESTs sequence with micro satellite; designing primer at two sides flank sequence of micro satellite repeat sequence; detecting optimization primer as the micro satellite label further; proceeding overall merit for micro satellite repeatability, stability, polymorphism; getting micro satellite label. The invention can be used for Pacific oyster species resource and genetic heterogeneity, molecular population genetics, species resource determination, genetic atlas construct, functional gene research, Pacific oyster breeding and cultivation.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
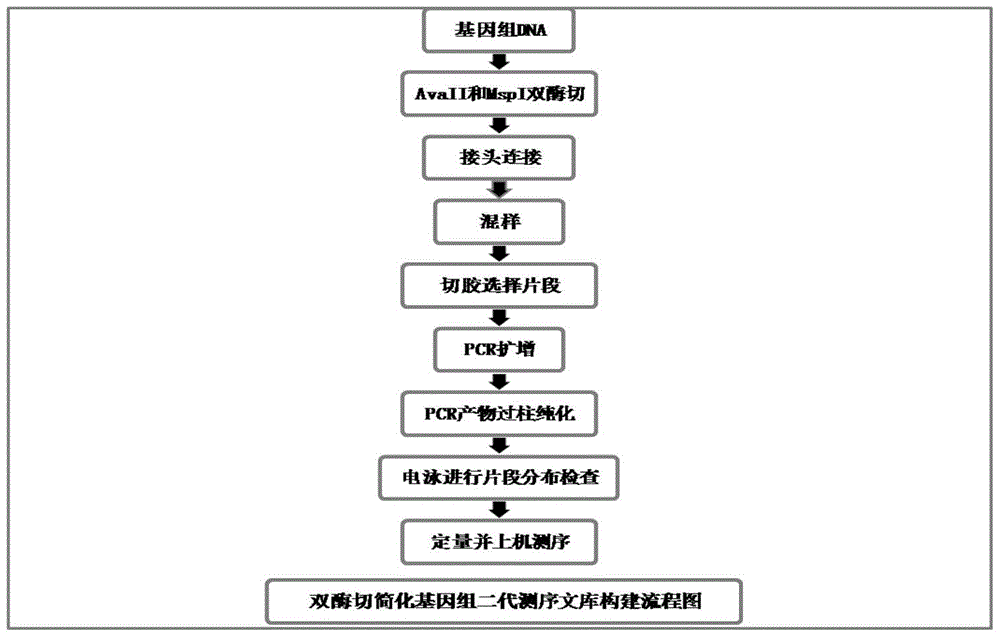
Construction method for double enzyme digestion simplified genome next generation sequencing library and matched kit
InactiveCN105696088AGet rid of dependenceSimplify the library building processMicrobiological testing/measurementLibrary creationGenomic sequencingEnzyme digestion
The invention provides a construction method for a simplified genome next generation sequencing library based on double enzyme digestion and a kit. Aiming at defects of an existing construction method for the double enzyme digestion simplified genome next generation sequencing library, the double enzyme digestion combined range is expanded, and excessive dependence on expensive instruments of constructing the simplified genome library is reduced, the library construction flow path is simplified, library construction cost is reduced, the sequencing efficiency is improved, and meanwhile the technology is easy and flexible to operate and easier for researchers to master and can be realized in a common molecule lab. The construction method is particularly suitable for miniature or medium-scale labs needing to conduct SNP molecular marker development, genetic map construction, population genetics research, phylogeny biological research and the like on a great number of species with incomplete reference genomes. The construction method has good practical application value and application prospects in the fields of molecular breeding of agriculture, conservation biology and evolutionary biology.
Owner:KUNMING INST OF BOTANY - CHINESE ACAD OF SCI
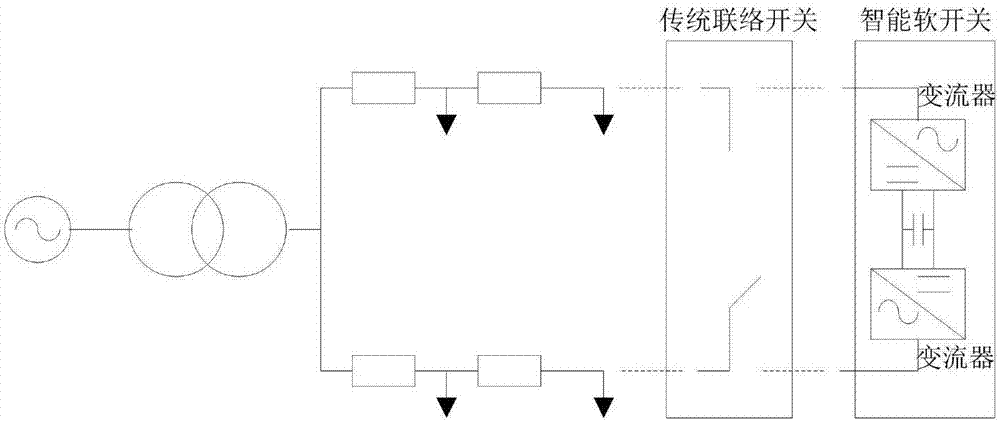
Computing method for distributed power maximum access capacity in flexible power distribution network based on SNOP
ActiveCN107196297ASolving high-dimensional nonlinear optimization problemsSolve the problem of easy to fall into local convergenceAc network circuit arrangementsGenetic algorithmDistributed power
The invention relates to a computing method for distributed power maximum access capacity in a flexible power distribution network based on SNOP. The computing method comprises the following steps: establishing a DG extremity access capacity model, using a network tide, feeder capacity and node voltage, distributed power single-point maximum access capacity and SNOP power as constraint conditions, selecting DG access capacity in maximum FDN as an optimal object, and adopting various population genetic algorithms to solve the model. According to the computing method, the distributed power maximum access capacity at an appointed position can be computed, and the optimal distributed power access position and capacity combination in the whole network can also be obtained in a planning angle.
Owner:STATE GRID TIANJIN ELECTRIC POWER +1
Detecting method for Litopenaeus vannamei Boone LvE165 microsatellite DNA marker
InactiveCN102146460AEasy identificationEasy to detectMicrobiological testing/measurementGenetic diversityMicrosatellite
The invention belongs to the field of molecular biology DNA marking technology and application, in particular to a detecting method for a Litopenaeus vannamei Boone LvE165 microsatellite DNA marker. The invention provides a Litopenaeus vannamei Boone LvE165 microsatellite specific DNA primer, a PCR (Polymerase Chain Reaction) system utilizing the primer and a detecting method for the Litopenaeus vannamei Boone LvE165 microsatellite DNA marker. The method comprises the following steps of: extracting a Litopenaeus vannamei Boone genome and diluting for later use; designing specific primers at both ends of a sequence by utilizing a Litopenaeus vannamei Boone LvE165 microsatellite DNA core sequence; and then carrying out PCR amplification on genome DNA of different groups of Litopenaeus vannamei Boone or individuals in the group by using the primer, analyzing a product and confirming the genotype of each individual so as to obtain a polymorphism genetic variation map. The invention is mainly applied to Litopenaeus vannamei Boone germplasm resource and genetic diversity analysis, molecular population genetics, construction of the genetic map, and the like.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Two SNP markers in close linkage with cucumber powdery mildew resistance and application thereof
ActiveCN104560983ASpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationGenotypeGenetic engineering
The invention relates to molecular markers in close linkage with cucumber powdery mildew resistance in the technical field of genetic engineering and provides SNP markers in close linkage with cucumber powdery mildew resistance QTL. The SNP markers are respectively named as Chr6SNP-pm01 and Chr6SNP-pm02. The Chr6SNP-pm01 is positioned at the loci 4, 450 and 551 of the 6th chromosome, and the Chr6SNP-pm02 is positioned at the loci 6, 822 and 412 of the 6th chromosome. The SNP markers in close linkage with cucumber powdery mildew resistance are screened by utilizing variant groups and population genetics, the cucumber genotypes are detected by virtue of methods for sequencing and transforming into dCAPS markers, and the difference of the cucumber powdery mildew resistance can be judged according to the genotypes, so that the breeding progress of the cucumber powdery mildew resistance strain is accelerated. Meanwhile, the two SNP markers lay a foundation for cloning cucumber powdery mildew resistance genes.
Owner:YANGZHOU UNIV
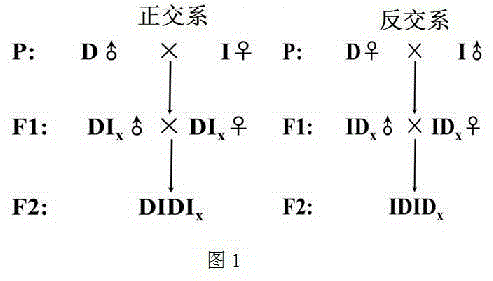
Establishment method of goat resource population
InactiveCN105123621AImprove economyImprove international competitivenessAnimal husbandryNational levelReciprocal cross
The invention belongs to the field of establishment of a population genetic resource bank, and provides a scientific establishment method of a goat resource population bank, and provides a goat reference family system establishment case, and relates to two national-level genetic resource varieties, namely, a Dazu black goat and an Inner Mongolia Cashmere goat (Alxa type). According to the resource population establishment method, forward cross and reciprocal cross of Dazu black goat and Inner Mongolia Cashmere goat (Alxa type) are utilized, random mating of genetic relationship is avoided for F1, so that Mendel genetic characters are separated, a goat economic character QTL or a functional gene is located by virtue of a genome means, and a relatively-precise genetic mark is provided for the goat breeding. The genetic resource bank comprises goat population, a genealogy inforamtion database, a phenotypic character database, a DNA sample library, an RNA sample library and a genetic variation information database.
Owner:SOUTHWEST UNIVERSITY
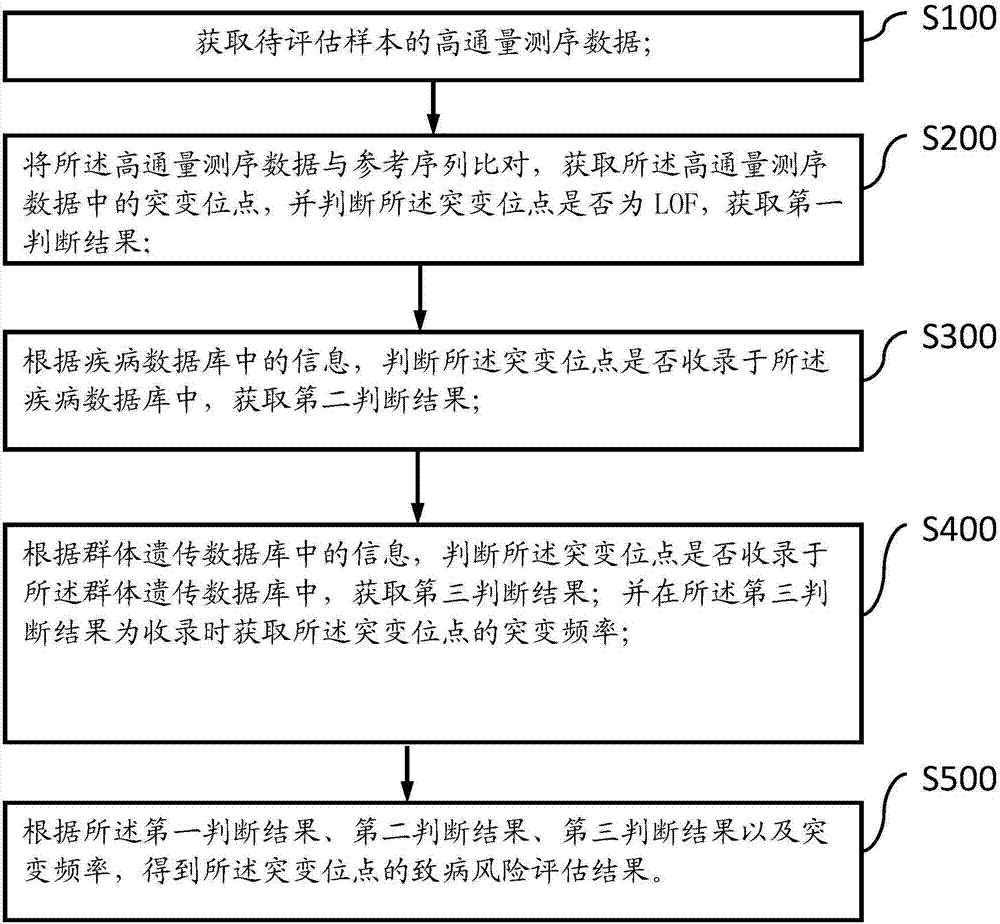
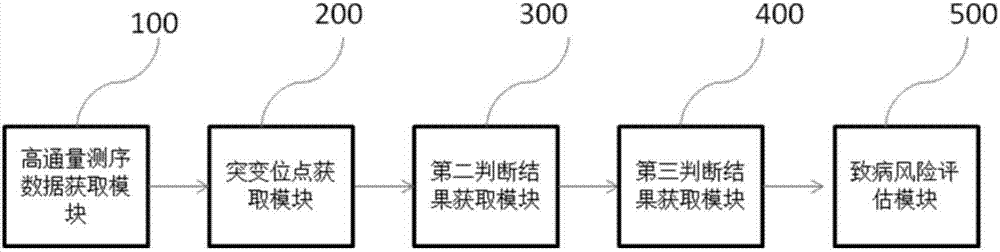
Gene mutation evaluation method and system
ActiveCN107229841ASimple and fast operationEasy to operateSequence analysisSpecial data processing applicationsPopulation geneticsSequencing data
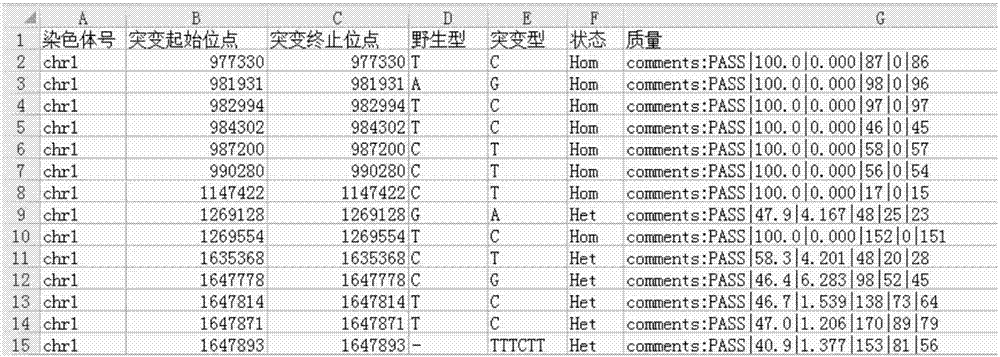
The invention discloses a gene mutation evaluation method. The gene mutation evaluation method comprises the following steps of obtaining high-throughput sequencing data of a sample to be evaluated; comparing the high-throughput sequencing data with a reference sequence, obtaining a mutant site in the high-throughput sequencing data, judging whether the mutant site is an LOF or not, and obtaining a first judgment result; judging whether the mutant site is recorded in the disease database or not according to information in the disease database, and obtaining a second judgment result; judging whether the mutant site is recorded in a population genetics database or not according to information in the population genetics database, and obtaining a third judgment result; obtaining a mutation frequency of the mutant site when the third judgment result is that the mutant site is recorded in the population genetics database; obtaining a pathogenic risk evaluation result according to the first judgment result, the second judgment result, the third judgment result and the mutation frequency. The invention further discloses a gene mutation evaluation system. The gene mutation evaluation method and system have the advantages of being easy to operate, efficient and high in applicability and saving time.
Owner:重庆金域医学检验所有限公司
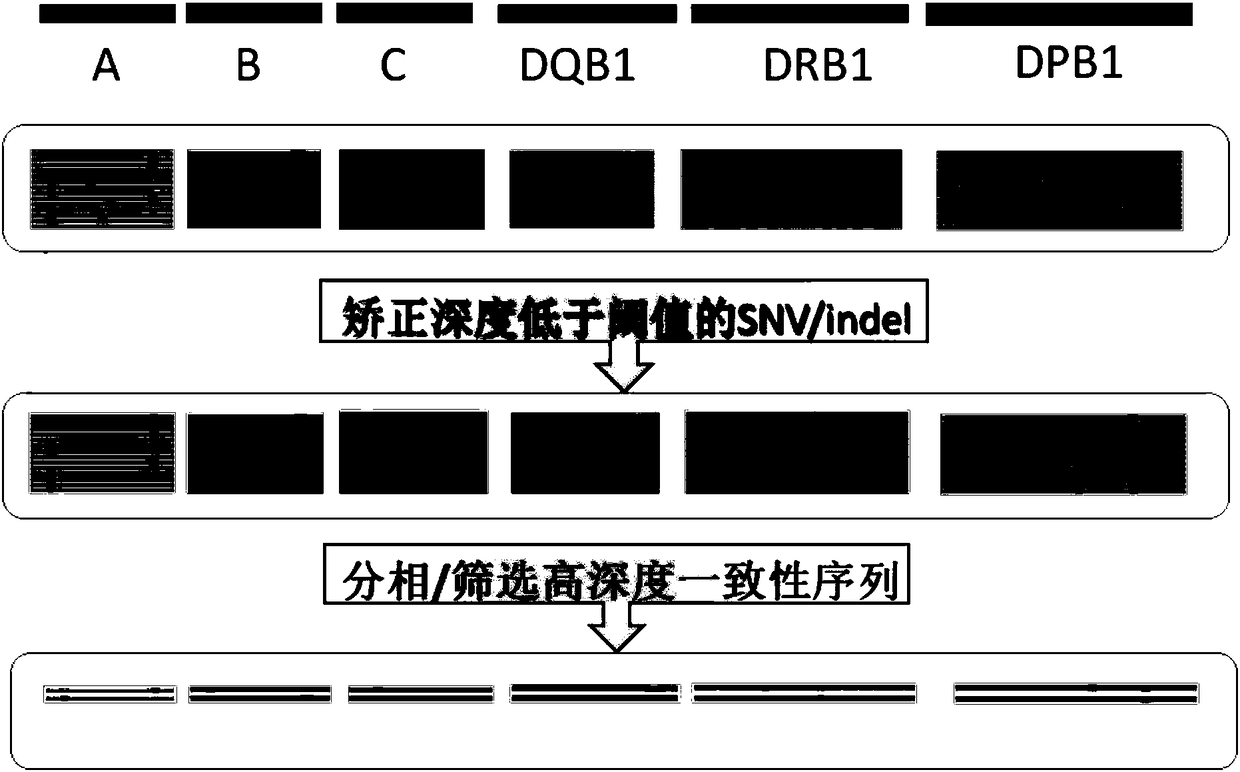
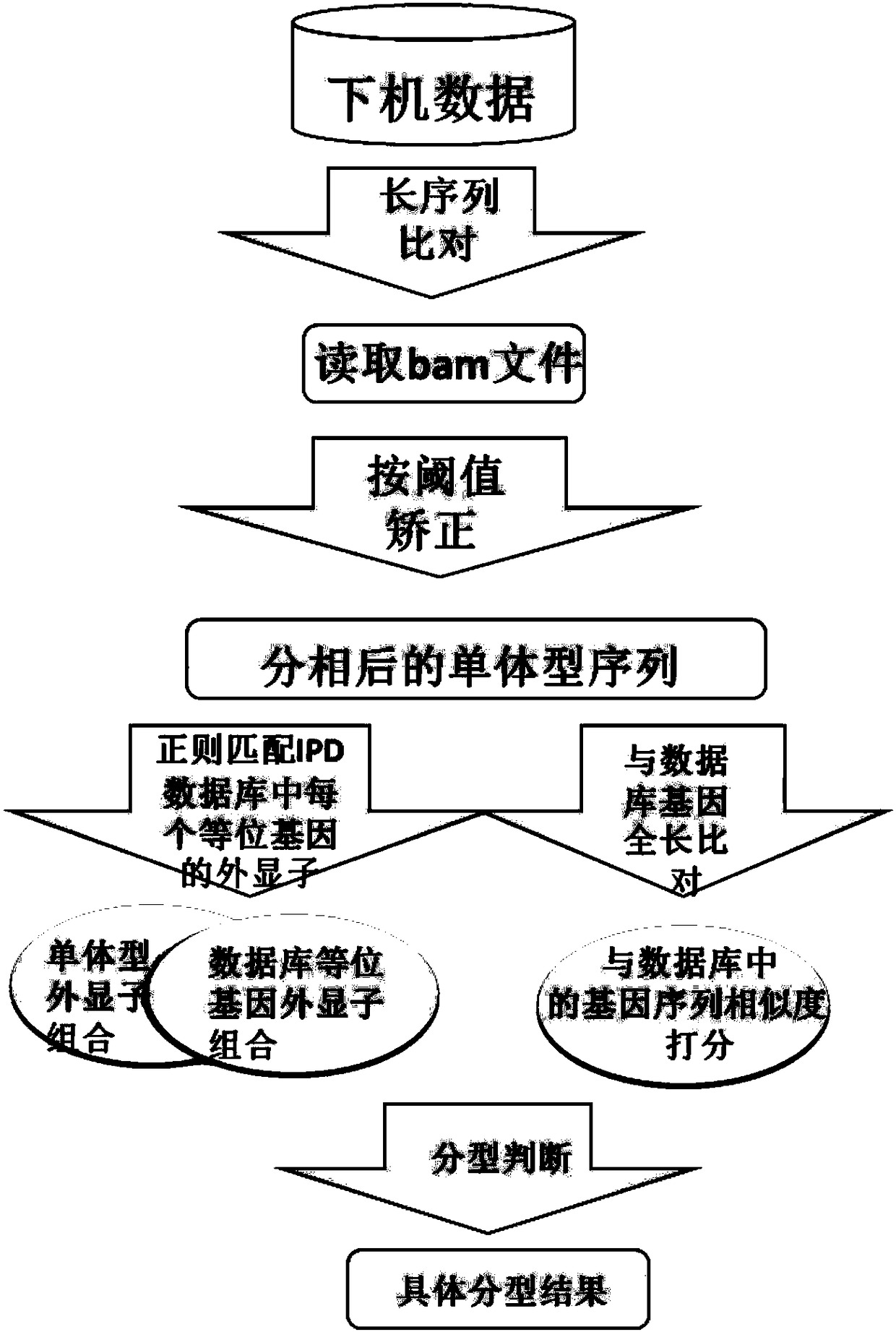
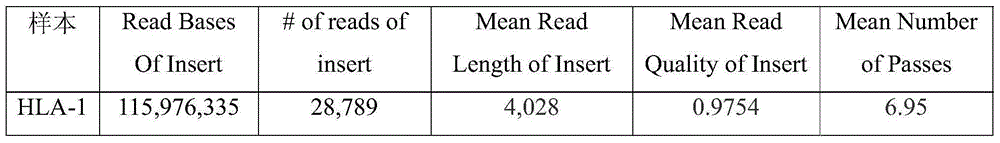
Three-generation sequencing platform-based HLA genotyping method
ActiveCN108460246AHigh resolutionDiversity revealsMicrobiological testing/measurementSequence analysisPopulation geneticsImage resolution
The invention discloses a three-generation sequencing platform-based HLA genotyping method. The method comprises the following steps of (1) performing PCR amplification on HLA genes needed to be genotyped; (2) after a PCR product is detected to be qualified, performing three-generation sequencing to obtain original data; (3) performing long sequence comparison on the original data and a referencegene sequence; (4) after comparison, correcting sequencing errors; (5) performing phase splitting to obtain a haplotype sequence; and (6) performing genotyping judgment. Compared with an existing HLAgenotyping method, the HLA genotyping method has the advantages of few requirements on computing resources, quick genotyping and high resolution, and has important values for application and basic research work of clinical transplantation tissue type matching, population genetics, anthropology, evolutiology and the like.
Owner:BEIJING GRANDOMICS BIOTECH
Litopenaeus vannamei osmotic regulation related functional gene EST-SSR markers and specific primers and detection method thereof
ActiveCN105969873AMicrobiological testing/measurementDNA/RNA fragmentationStructure analysisMicrosatellite
The invention discloses litopenaeus vannamei osmotic regulation related functional gene EST-SSR markers and specific primers and a detection method thereof. Litopenaeus vannamei genome DNA serves as a template, the specific primers of the litopenaeus vannamei osmotic regulation related functional gene EST-SSR markers are used for conducting PCR amplification on microsatellite locus sequences, and then typing is conducted on PCR amplification products through a sequenator. The specific primers and detection method of the litopenaeus vannamei osmotic regulation related functional gene EST-SSR markers can be used for population genetic structure analysis and molecular marker assisted breeding of litopenaeus vannamei and particularly have an important application value in breeding of good salt-tolerant varieties of the litopenaeus vannamei.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Method for rapidly detecting microsatellite markers of Charybdis feriatus
InactiveCN103305611BFast detection methodThe detection method is accurateMicrobiological testing/measurementGenomic DNAGenotype
The invention relates to a method for rapidly detecting microsatellite markers of Charybdis feriatus. The method comprises the following steps of: (1) extracting a genomic DNA (Deoxyribose Nucleic Acid) of the Charybdis feriatus; (2) obtaining a gene sequence containing microsatellite repeats according to the functional gene sequence of the Charybdis feriatus in a GeneBank database; (3) designing a microsatellite marker primer; (4) implementing PCR (Polymerase Chain Reaction) amplification on the genomic DNA of different individuals of the Charybdis feriatus; and (5) detecting denaturing polyacrylamide gel electrophoresis of PCR products. The method has the advantages of rapidness, accuracy, sensitivity and the like, and can intuitively detect the genotype of different individuals of the Charybdis feriatus, so that a polymorphism map of genetic variation of the Charybdis feriatus on a functional gene microsatellite site is obtained quickly; the microsatellite markers can be applied to the fields of analysis on genetic variation and population genetic diversity of the Charybdis feriatus.
Owner:EAST CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Reagent kit for extracting DNA in Chinese alligator chorion film and use method thereof
InactiveCN101368179AHigh purityIncrease concentrationMicrobiological testing/measurementDNA preparationHigh concentrationChinese alligator
The invention discloses a kit for extracting DNA in the egg shell membrane of Yangtze alligator and a use method thereof, and the kit comprises the following reagents: a reagent A, a reagent B, a reagent C, a reagent D, a reagent E and a reagent F. The template DNA which is extracted by the kit has good purity, and the kit can be used for gene amplification sequencing and typing. In the amplification of the 12S rRNA gene, the mitochondrial D-loop region and the Cyt b gene, all the extracted shell membrane DNA templates can be successfully amplified to obtain the fragment products with expected identical sizes, high concentration and high specificity. Microsatellite DNA marker specificity primers are utilized to obtain typing data through the PCR amplification to be used for the identification in disputed paternity of Yangtze alligator. In addition, the method also can be applied to the extraction of other ovipara such as the turtles, the birds and the like. The obtained template DNA can be widely applied to the studies of the ovipara on the fields such as population genetic analysis, molecular evolution, evolution of relationship among individuals and the like.
Owner:ANHUI NORMAL UNIV
High-efficiency screening method for microsatellite marker of Marbled Sand Goby and application of same
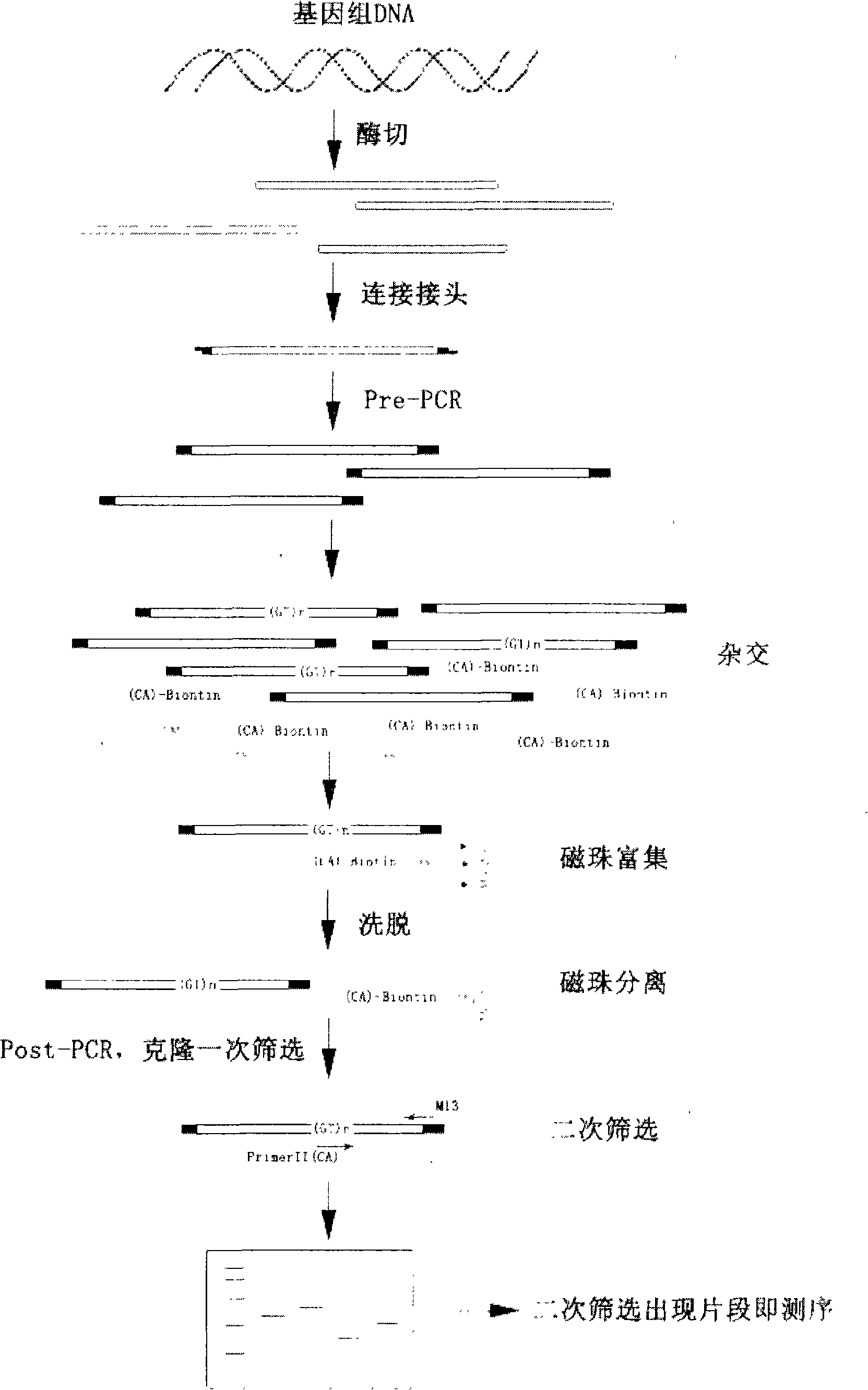
InactiveCN102134586ARapid development of germplasm resources protectionImprove screening efficiencyMicrobiological testing/measurementGerm plasmEnrichment methods
The invention belongs to the technical field of DNA markers of molecular genetics of fishes, and particularly relates to the fast and highly efficient screening and application of the microsatellite loci of the Marbled Sand Goby. The method comprises the following steps: large mass DNA enzyme ligation, connection; low cycle index Pre-PCR; construction of the enriched library of microsatellite fragments by utilizing a biotinylated probe and streptomycin magnetic beads; screening of positive clones, and further improvement of the detection efficiency of the microsatellite loci through secondary screening; and population genetic analysis by using an obtained microsatellite primer. The invention improves the existing magnetic-bead enrichment method, efficiently and quickly screens the microsatellite loci, reduces the sequencing cost, and simultaneously overcomes the deficiency on safety during secondary screening adopting isotope. The obtained microsatellite marker provides scientific basis and detection means for the germ plasm resource protection, the germ plasm improvement, the genetic breeding, and the like of the Marbled Sand Goby.
Owner:HAINAN UNIVERSITY
Family breeding method for pearl white strain red tilapia
ActiveCN104430084AQuick filterBreeding traits intuitiveClimate change adaptationPisciculture and aquariaJuvenile fishTilapia
The invention provides a family breeding method for pearl white strain red tilapia. The method includes the steps that (1) parent fish selection is conducted, the red tilapia with the pearl white body surface and the whole body being free of red spots or black spots is picked out from a red tilapia group with serious body color differentiation, and the red tilapia with obvious growth advantages is used as standby parents; (2) family establishment is conducted, the selected male and female parents are placed into a net cage for carrying out adaptation reinforcement cultivation, and manual orientation mating, hatching and seedling cultivating are independently performed according to the proportion of one to one; (3) pearl white strain breeding of the red tilapia is conduced, the growing juvenile fish stage and the adult fish stage of the red tilapia are determined, the pearl white red tilapia family with the high proportion and good growth traits are screened out to be used as the breeding base family of the next generation, 2-4 generation breeding is conducted according to the method, and accordingly the pearl white strain red tilapia can be bred. By the adoption of the method, inbreeding is avoided, the variety degeneration of the red tilapia is effectively prevented, and the population genetic diversity is kept; the breeding process is accelerated, and the breeding efficiency is improved.
Owner:FRESHWATER FISHERIES RES CENT OF CHINESE ACAD OF FISHERY SCI
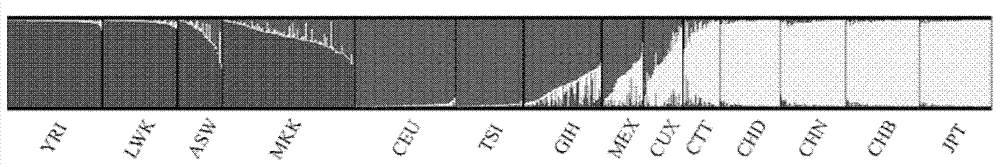
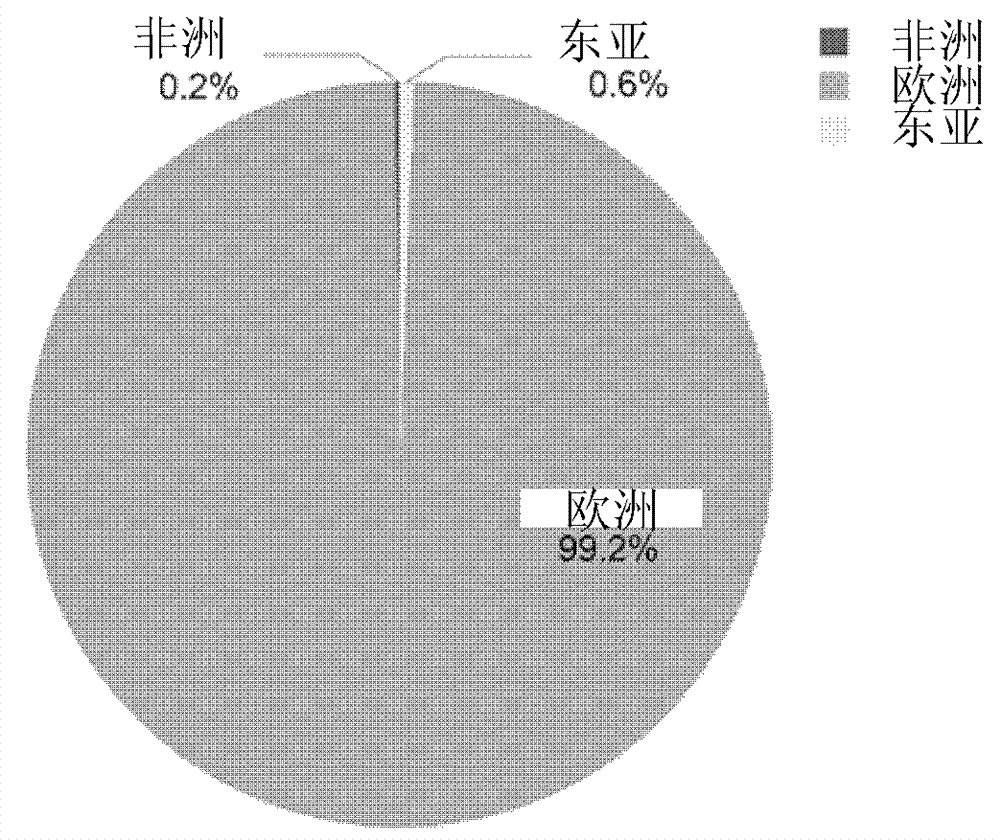
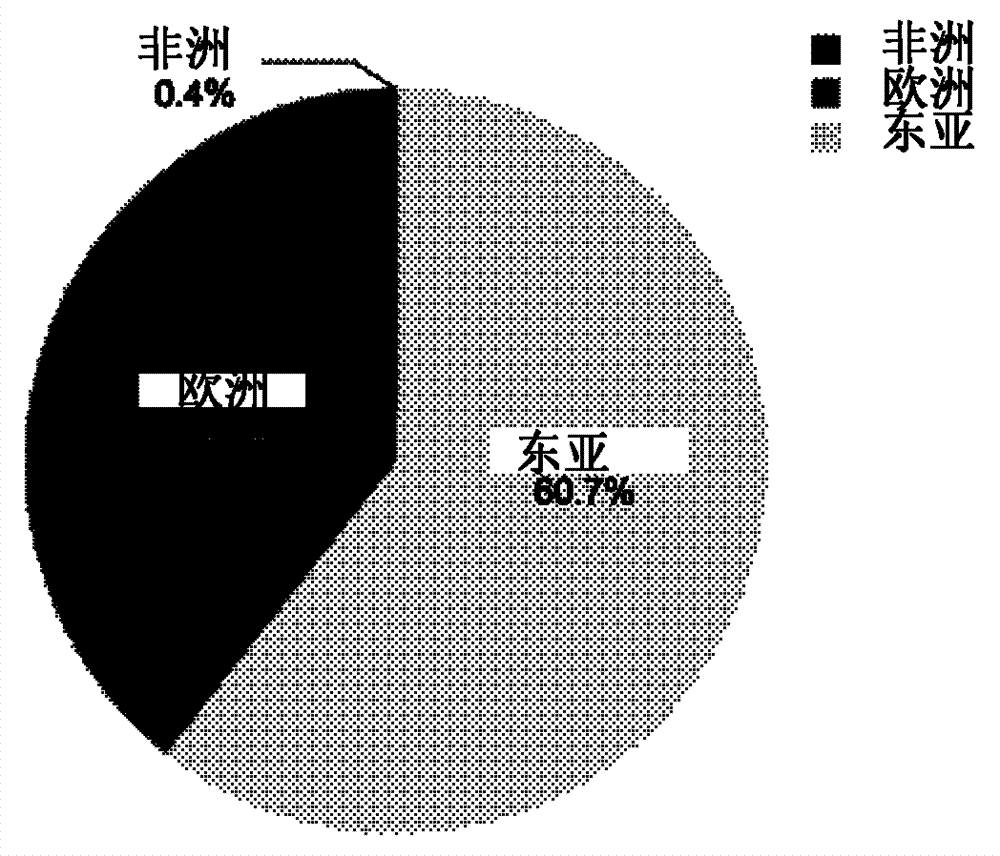
Method and system for performing African, European and East Asian population genetic principal component analysis to unknown-source individual
ActiveCN104212886AClarify the direction of investigationLess templatesMicrobiological testing/measurementDNA/RNA fragmentationPopulation geneticsAsian population
The invention provides a method and a system for performing African, European and East Asian population genetic principal component analysis to an unknown-source individual. The method includes following steps: extracting DNA from the unknown-source individual; obtain 27 genotypes of specific locus of African, European and East Asian populations; and obtaining a population genetic principal component analysis result of the genotypes of the specific locus through STRUCTURE software. By means of the scheme in the invention, the African, European and East Asian population genetic principal component analysis of the unknown-source individual can be achieved and data support for inferring African, European and East Asian population sources of the unknown-source individual is provided.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
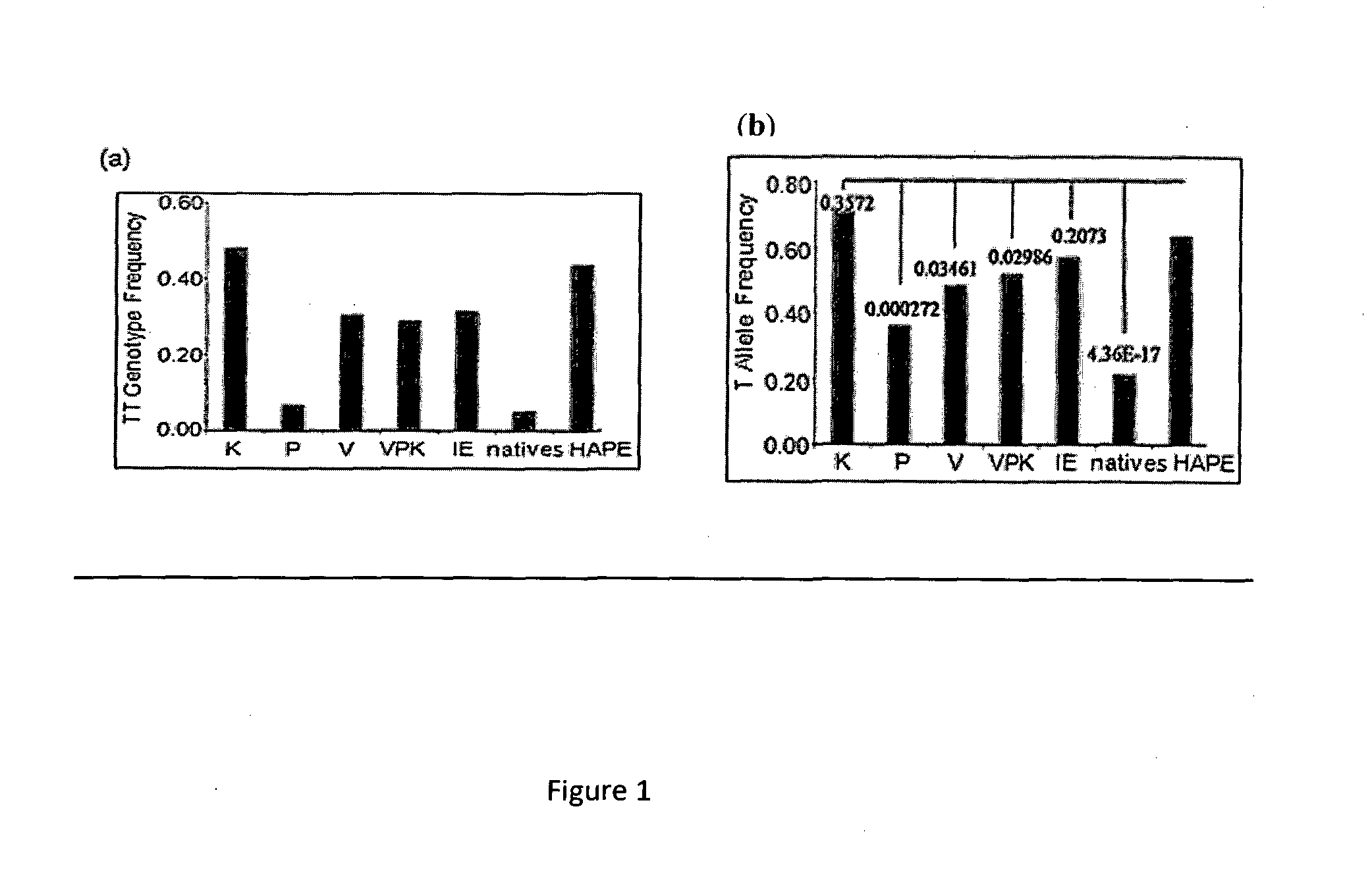
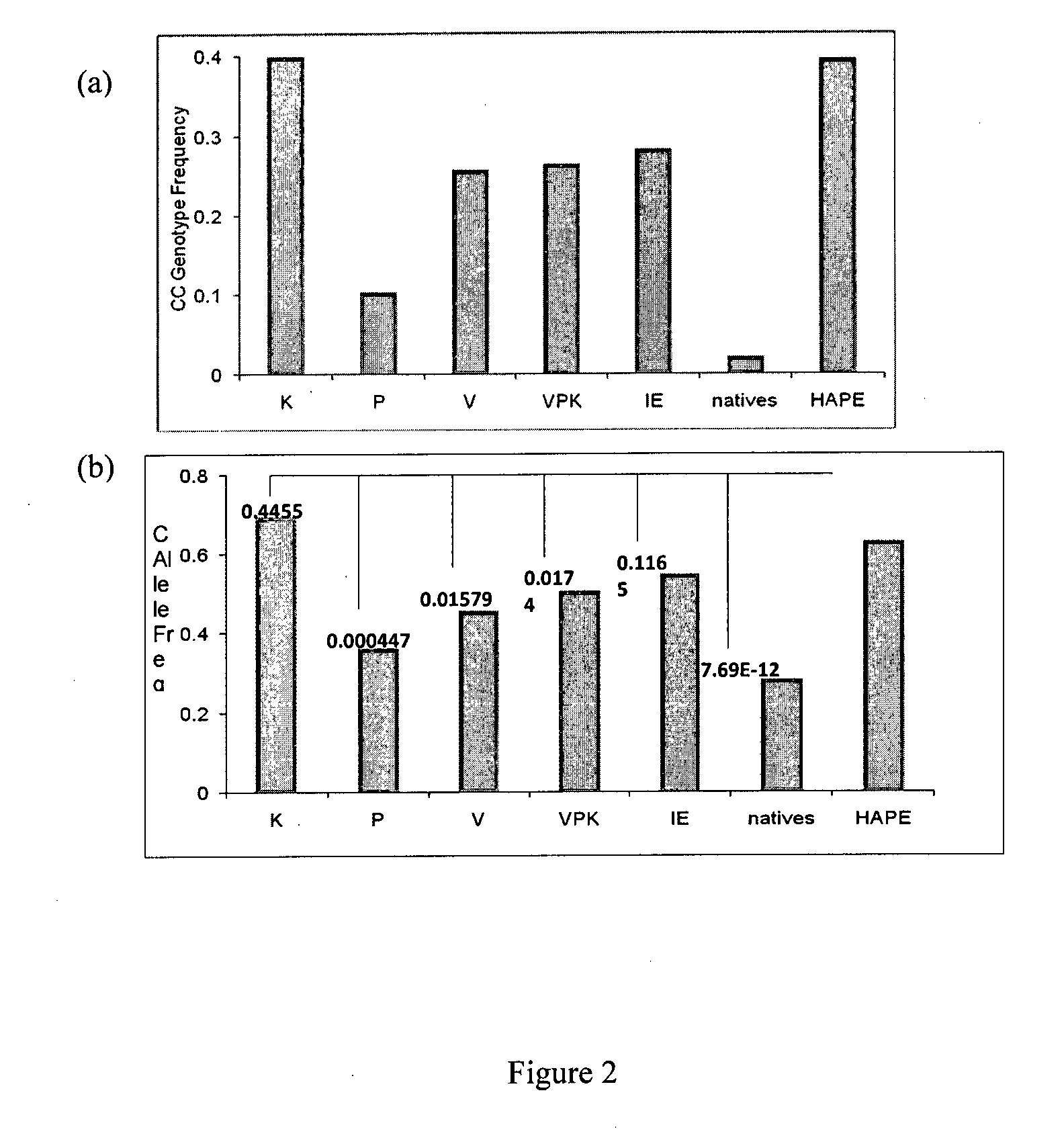
Biomarker for Detecting High-Altitude Adaptation and High-Altitude Pulmonary Edema
InactiveUS20140030709A1Reduce riskSugar derivativesMicrobiological testing/measurementProlyl HydroxylasesBiomarker (petroleum)
Owner:COUNCIL OF SCI & IND RES
Cucumber fruit beta carotene content related SNP marker and its application
InactiveCN103146659ASpeed up the breeding processReliable resultsMicrobiological testing/measurementOxidoreductasesBeta-CarotenePhenotypic trait
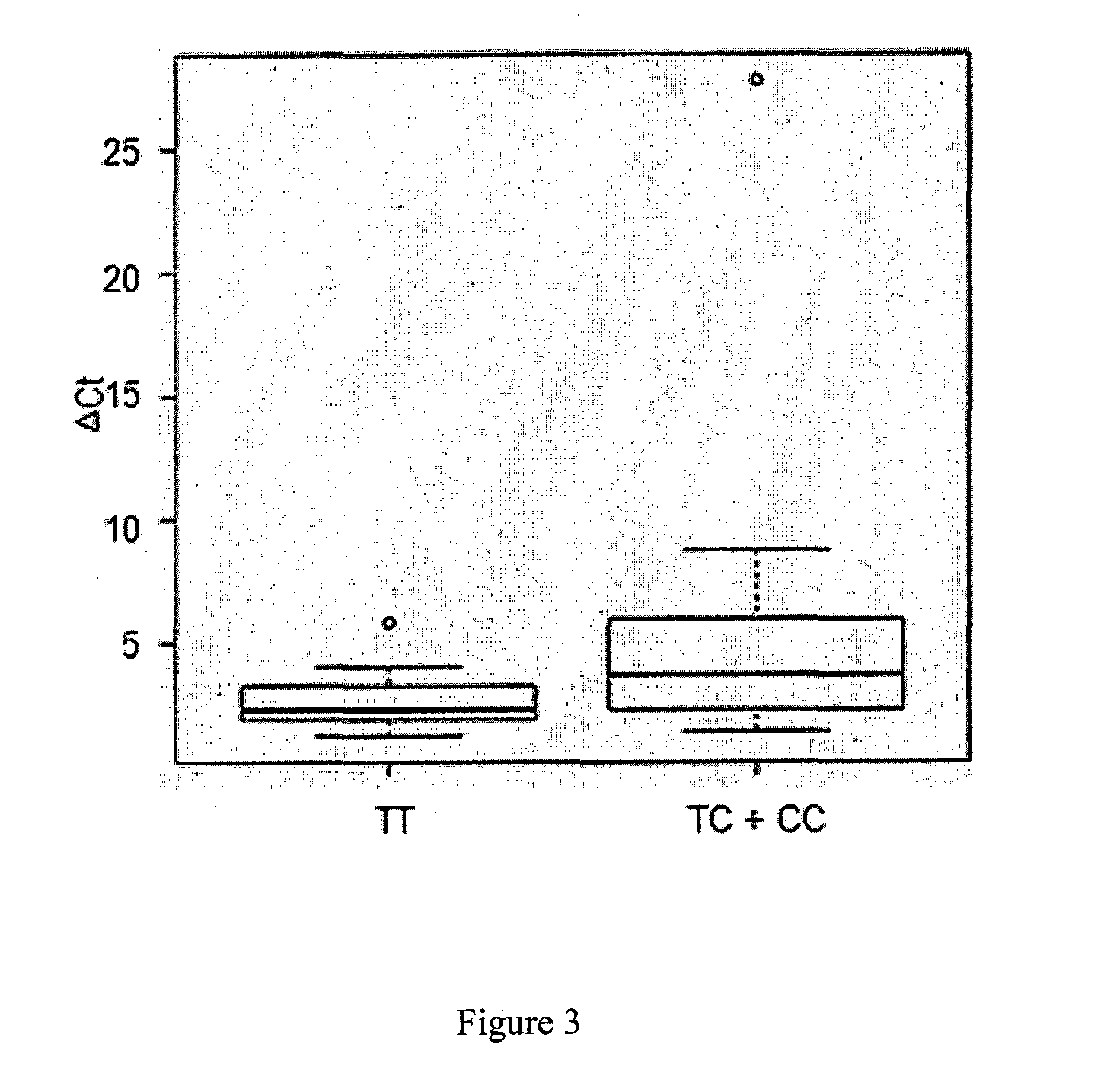
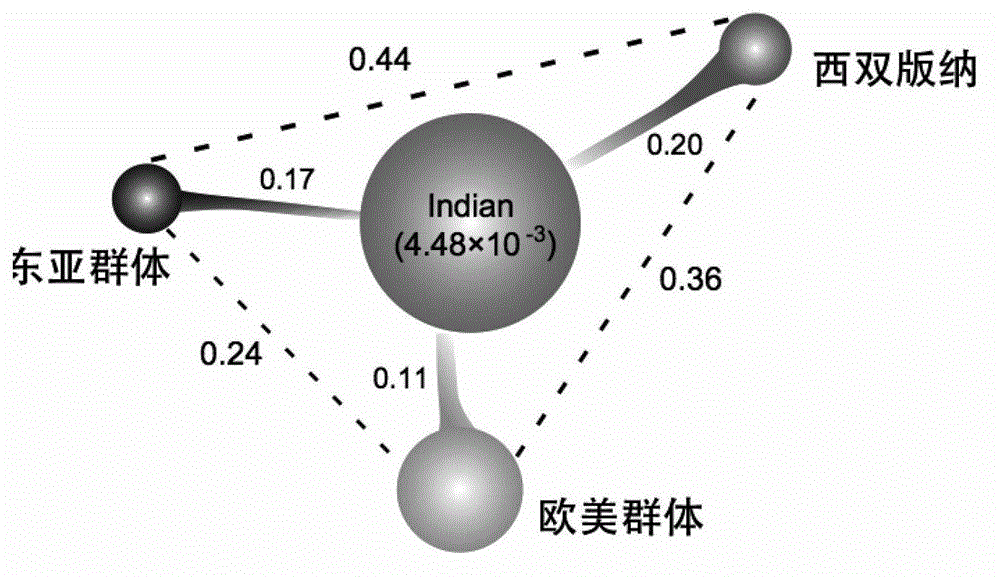
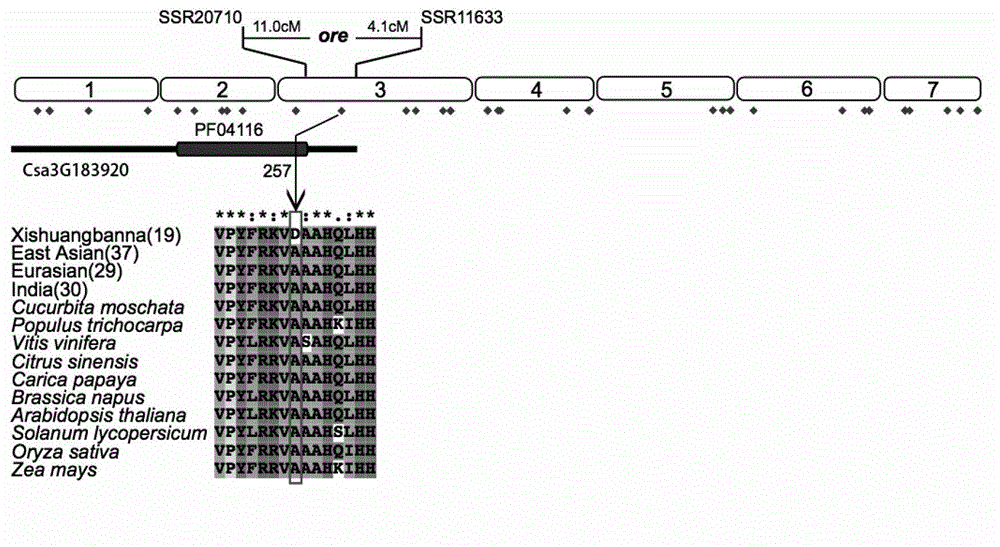
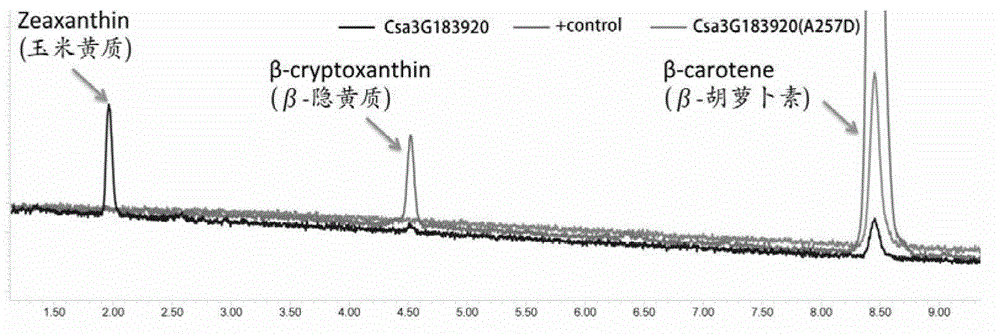
The invention provides a cucumber fruit beta carotene content related SNP marker and its application. The SNP marker is located at the 770bp site in a gene sequence of a coded cucumber Csa3G183920 protein, the cucumber fruit beta carotene content is high at the site with A as a basic group, and the flesh color is orange yellow. The cucumber fruit beta carotene content is low at the site with C as a basic group, and the flesh color is white. In the invention, variomics, population genetics and other methods are utilized to screen the SNP marker related to cucumber fruit beta carotene content and flesh color characters. And through a biochemical method, the molecular mechanism that Csa3G183920 regulates fruit color by degrading beta carotene is illuminated. The genotype of a to-be-tested cucumber can be detected through sequencing, dCAPS or other methods. And according to the genotype, the cucumber fruit beta carotene content and the flesh color can be determined, thus accelerating the cultivation process of high nutrition cucumber varieties.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Detection method of turbot FF0901 microsatellite marker by utilizing specific primers
InactiveCN101705295AEasy to detectEasy accessMicrobiological testing/measurementNegative strandGenetics
The invention discloses a detection method of a turbot FF0901 microsatellite marker by utilizing specific primers, comprising the following steps: firstly extracting turbot genomic DNA; then utilizing a sequence containing a microsatellite in an enrichment library of the turbot microsatellite; designing specific primers at two ends of the sequence as a positive strand 5'-TTACGCTTCGCATCGCTCAT-3', and a negative strand 5'-TGCCAGGCCACCAGACG-3'; carrying out PCR amplification on the turbot genomic DNA; separating the obtained PCR product through modified polyacrylamide gel electrophoresis; analyzing strips generated on the product to obtain a turbot genetic polymorphism map. The method of the invention is suitable for the detection technologies of turbot population genetic marker, genealogy authentication, genetic linkage map construction and the like; the PCR amplification product presents better polymorphism in turbot population detection; and the method of the invention can rapidly acquire a genetic variation map of the turbot FF0901 microsatellite marker, and conveniently, rapidly, accurately and intuitively detect each individual genotype of the turbot.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
HLA (human leucocyte antigen) typing method based on PacBio RS II sequencing platform
The invention discloses an HLA (human leucocyte antigen) typing method based on a PacBio RS II sequencing platform. A collected sample is subjected to DNA extraction and then to PCR (polymerase chain reaction) amplification, PCR products are mixed to establish a 10k library, and PacBio RS II sequencing is performed; then, original data obtained by sequencing are corrected, and HLA typing is performed with software programs. Compared with existing HLA typing methods, the HLA typing method has super-high resolution and is of important value to applications such as clinical graft tissue matching, population genetics, anthropology and evolutiology, as well as basic research work.
Owner:BEIJING GRANDOMICS BIOTECH
Primer combination and kit for simultaneous detection of 93 bovine genetic defect genes and lethal haplotypes
ActiveCN107099607AAccurate amplificationImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceAssortative mating
The invention discloses a primer combination and a kit for simultaneous detection of 93 bovine genetic defect genes and lethal haplotypes. Through use of the primer combination and the kit, casual mutation sites (SNP and insertion or deletion of short fragments) of the 93 genetic defect genes (such as PIRM syndrome, crooked tail syndrome, epidermolysis bullosa and the like) and the lethal haplotypes (HH1, HH3, HH4, HH5 and JH1) can be detected in one time, cattle with monogenic inheritance defect gene carriers and lethal haplotype carriers can be screened, and accurate guidance is provided for cattle breeding and genetic evaluation, herd selection and assortative mating, population genetic improvement, cultivation of new varieties and genetic resource protection. The primer combination and the kit have the characteristics of high throughput, low cost, high accuracy, and the like, is suitable for the varieties of beef cattle and dairy cattle, and can be widely used in the breeding and reproduction field of the cattle.
Owner:DAIRY CATTLE RES CENT SHANDONG ACADEMY OF AGRI SCI
Method for identifying odontobulis mpotamophila family
InactiveCN105331732AGuaranteed accuracyReduce mistakesMicrobiological testing/measurementDNA paternity testingGermplasm
The invention relates to the technical field of fish molecular marker and genetic resource identification, in particular to a method for identifying an odontobulis mpotamophila family. The method comprises the steps of 1, building and breeding an odontobulis mpotamophila family to be tested; 2, extracting DNAs of parents and hybrids of the odontobulis mpotamophila family to be tested; 3, screening and synthetizing polymorphism microsatellite primers of odontobulis mpotamophila; 4, amplifying microsatellite primers of the parents and hybrids of odontobulis mpotamophila; 5, identifying systems and genetic relationships of the odontobulis mpotamophila. According to the method, different systems can be accurately and rapidly identified, paternity testing can be performed on the odontobulis mpotamophila, artificial breeding is optimized, in-breeding degeneration is avoided, technical support is provided for building the family tree of odontobulis mpotamophila cultured stocks and formulating scientific population genetic management strategy, and the reliable method is provided for later large-scale family identification and paternity testing of odontobulis mpotamophila.
Owner:NANJING NORMAL UNIVERSITY
Detection method for Apostichopus japonicas AjE101 micro-satellite DNA label
InactiveCN102140522AGood polymorphismImprove stabilityMicrobiological testing/measurementGerm plasmGenetic diversity
The invention belongs to the field of molecular biology DNA labeling technology and application, and in particular relates to Apostichopus japonicas express sequence tag EST micro-satellite label screening development and application. The invention provides Apostichopus japonicas AjE101 micro-satellite specific DNA primers, a polymerase chain reaction (PCR) reaction system by utilizing the primers, and a detection method for an Apostichopus japonicas AjE101 micro-satellite DNA label. The detection method comprises the following steps of: extracting genome of Litopenaeus vannamei Boone; designing the specific primers at two ends of an Apostichopus japonicas AjE101 micro-satellite DNA core sequence; and performing PCR amplification on genome DNA of different groups of the Litopenaeus vannamei Boone or individuals in the group by using the primers, analyzing products, and determining the genome of each individual so as to obtain an Apostichopus japonicas polymorphism genetic variation map. The invention is mainly applied to Apostichopus japonicas germ plasm resource and genetic diversity analysis, cular population genetics, construction of genetic maps and research of functional genes.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Screening method of scylla paramamosain SNPs molecular marker
InactiveCN103045739AShort experiment cycleLow costMicrobiological testing/measurementNucleotideScylla paramamosain
The invention relates to a screening method of a scylla paramamosain SNPs molecular marker, which comprises the following steps of: (1) obtaining a scylla paramamosain genome sequence through a genomic library or a GenBank database obtained by sequence determination; (2) carrying out primer design on the obtained genome sequence, carrying out sequence determination after PCR (Polymerase Chain Reaction) amplification, contrasting the sequences of different individuals and verifying candidate SNPs (Single Nucleotide Polymorphisms); and (3) designing a primer with a specific allete for a candidate SNPs site and carrying out verification and genetic typing on the SNPs by adopting a real-time fluorescent quantitative PCR dissolving curve method. The screening method has the advantages of short experimental period, low cost, simplicity in operation and the like, and the genetic types of the different individuals can be accurately detected to provide a novel genetic marker for scylla paramamosain genetic variation analysis, population genetic diversity evaluation and molecular marker-assisted breeding.
Owner:EAST CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Population genetic evolution map and construction method thereof
ActiveCN110910959AGenetic relationship is clearBuild accuratelySequence analysisSpecial data processing applicationsCold spotGenetics
The invention provides a population genetic evolution map and a construction method thereof. The construction method comprises the following steps: selecting a recombined cold spot region; constructing a phylogenetic tree by utilizing the SNP sites of the recombined cold spot region; performing clustering analysis on the phylogenetic tree, and determining an evolution relationship between speciesso as to obtain a population genetic evolution map. According to the method, the phylogenetic tree is constructed by selecting SNP sites of the recombined cold spot region (such as a centromere region); on one hand, due to low recombination occurrence rate and stable inheritance between parents and filial generations, the genetic evolution relationship is clear, the constructed phylogenetic tree is more accurate, and on the other hand, the number of SNP sites in the selected sequence is smaller than that of whole genome sequences and is more and more comprehensive than that of SNP sites in a haplotype analysis method. Therefore, the phylogenetic tree can be quickly and accurately constructed.
Owner:CHINA NAT RICE RES INST
QTL related to buffalo milk production traits as well as screening method and application of QTL
ActiveCN110289048AIncrease milk productionHigh milkfatProteomicsGenomicsQuantitative trait locusScreening method
The invention discloses QTL related to buffalo milk production traits and a screening method and application thereof. The screening method of the QTL related to buffalo milk production traits comprises the specific steps of sample collection and genotyping, population genetic analysis, selection signal identification and QTL identification related to buffalo milk production traits. The method specifically comprises: screening out four QTLs (Quantitative Trait Locus) containing the most significant SNP, such as 11_Block2, 12_Block2, 16_Block1 and 19_Block5; applying H1H3, H3H4 or H1H2 in 11-Block2, H2H2 in 12_Block2, H2H2 in 16_Block1, H1H4 in 16_Block1 and H2H4 in 19_Block5 to judge and breed buffalo with excellent milk production traits. Technical support is provided for breeding of buffalo milk with excellent milk production performance, and the method is a new method for auxiliary detection of buffalo milk production traits.
Owner:GUANGXI ZHUANG AUTONOMOUS REGION BUFFALO INST
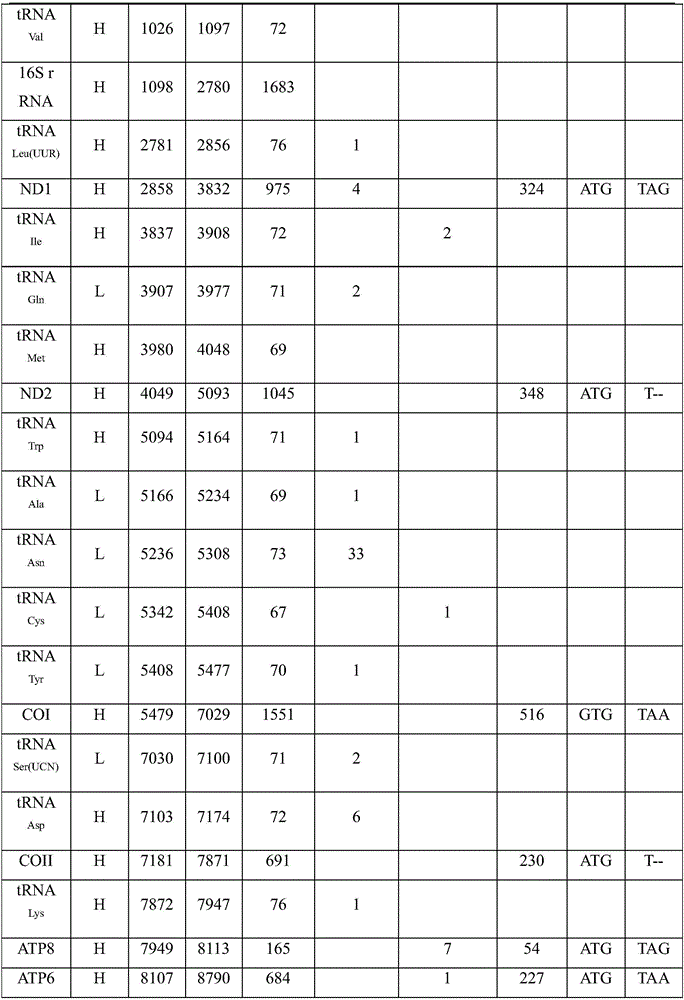
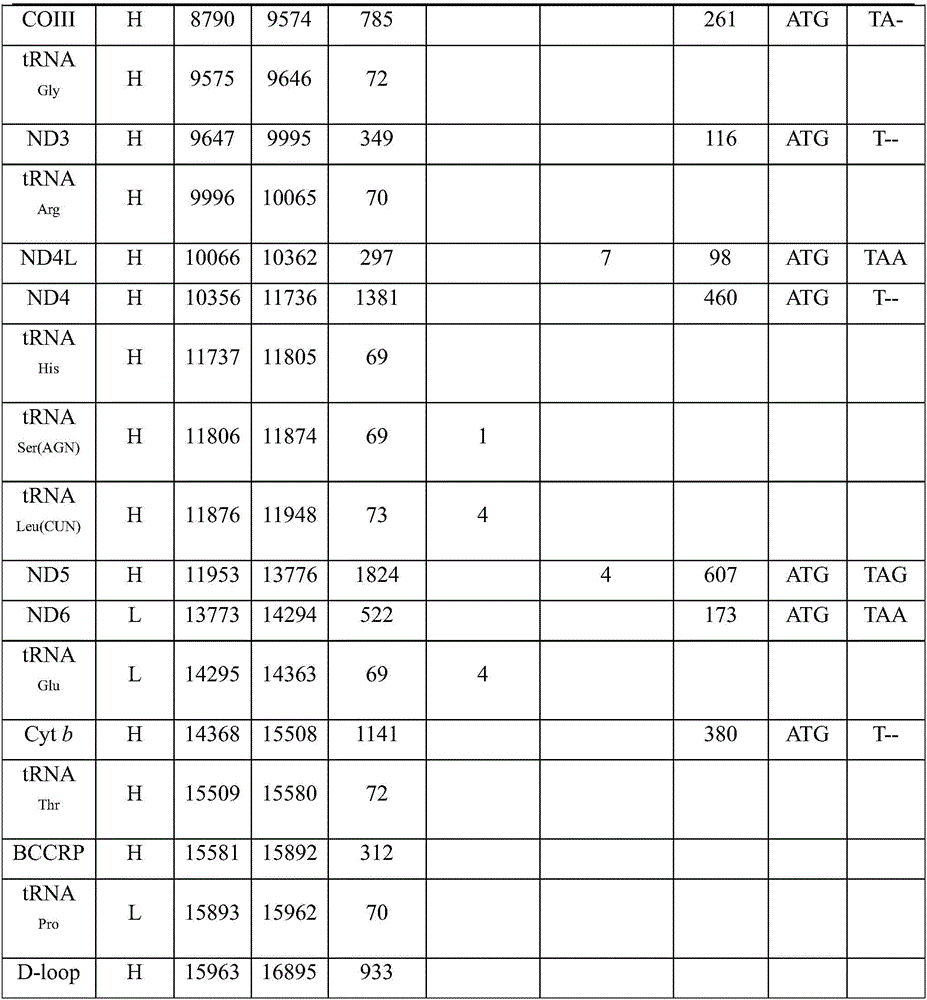
Complete sequence amplification method for mitochondrial genome of diptychus maculates steindachner
The invention discloses a complete sequence amplification method for a mitochondrial genome of diptychus maculates steindachner. The complete sequence amplification method specifically comprises the following steps: (1) amplifying four gene segments Cyt b, COI, ND4 and 16S rRNA of the mitochondrial genome of the diptychus maculates steindachner; and (2) carrying out LA-PCR amplification on the mitochondrial genome of the diptychus maculates steindachner. According to the complete sequence amplification method, the four gene segments Cyt b, COI, ND4 and 16S rRNA of the mitochondrial genome of the diptychus maculates steindachner are amplified, four pairs of LA-PCR primers are designed according to an obtained sequence and are used for carrying out large-segment sequence amplification, and 28 sequencing primers are utilized for carrying out PCR amplification on the other parts of the mitochondrial genome of the diptychus maculates steindachner by virtue of a primer PCR-Walking method so as to obtain a whole-genome sequence, with a length of 16895bp, of a mitochondrion of the diptychus maculates steindachner. The inheritance and evolution of the diptychus maculates steindachner are researched in a molecular level, and particularly mtDNA is used as a molecular marker, so that the basic data and technical support are provided for the researches of species identification, population genetic structures and system evolution of the diptychus maculates steindachner.
Owner:ZHEJIANG OCEAN UNIV
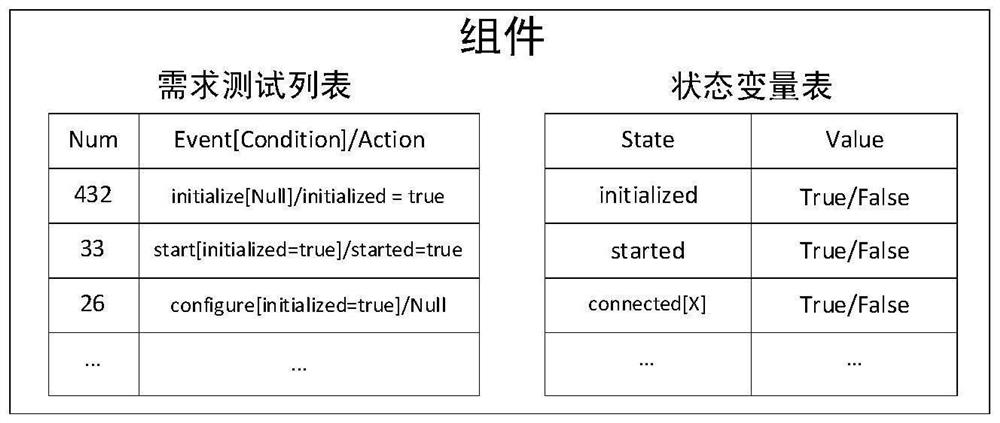
Software radio conformance test method based on multi-population genetic algorithm
ActiveCN111782544AEasy to manageAvoid false resultsSoftware testing/debuggingGenetic algorithmsTest efficiencySoftware define radio
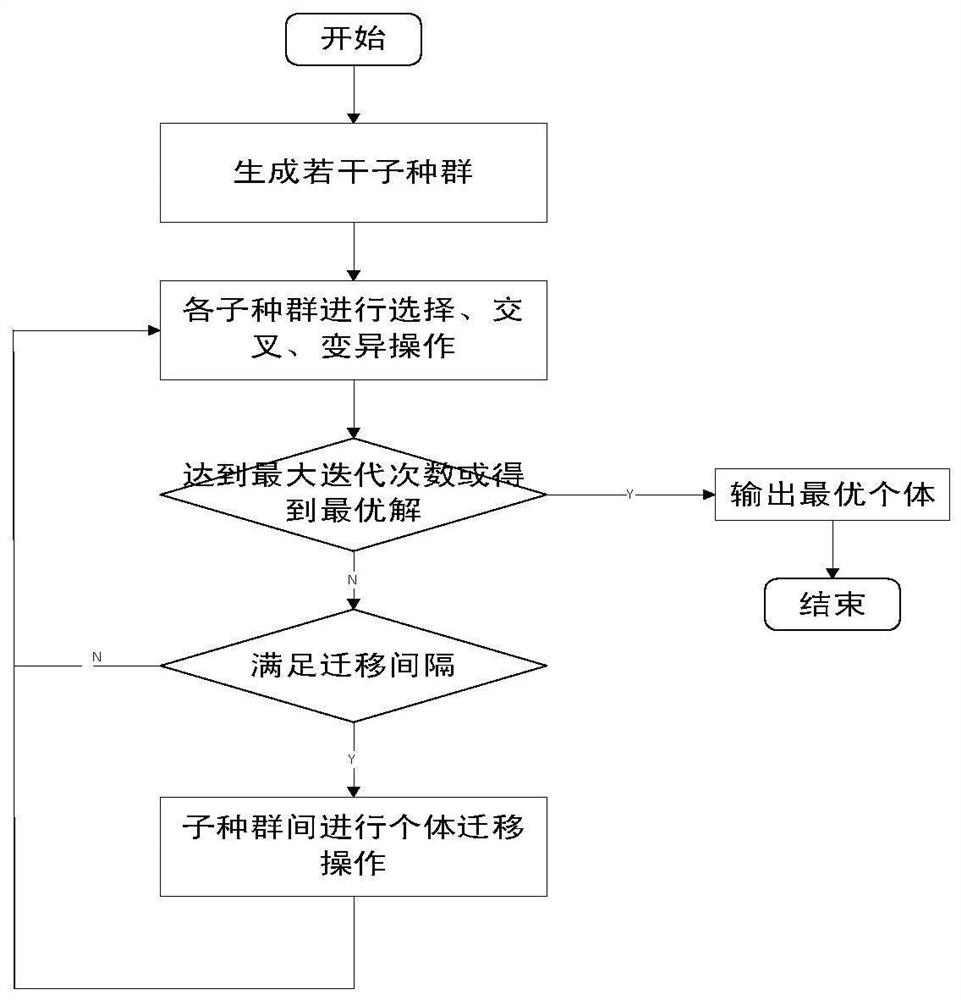
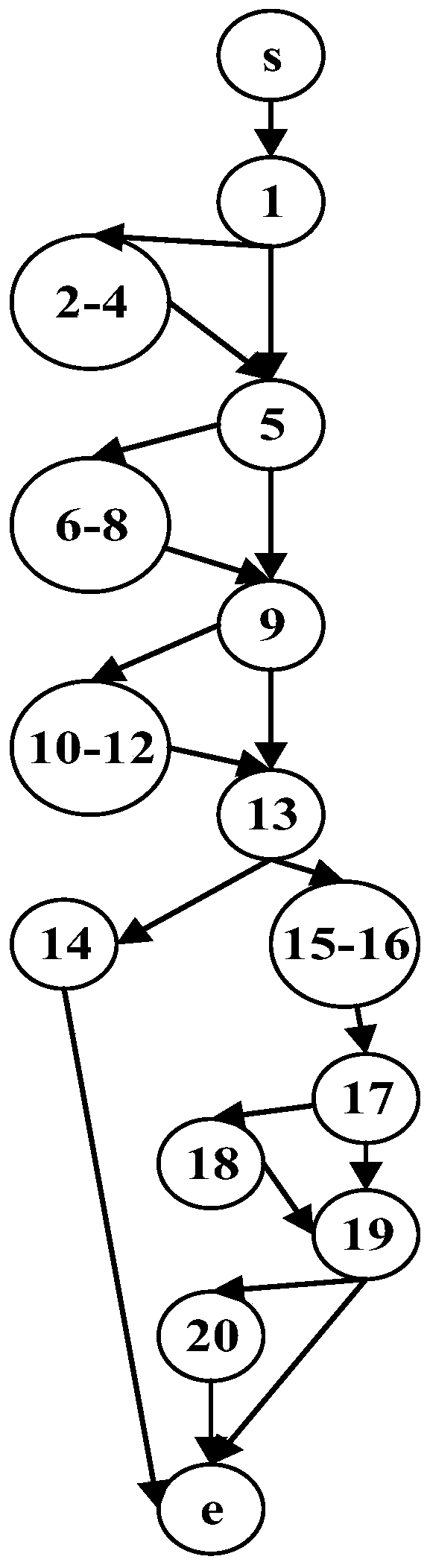
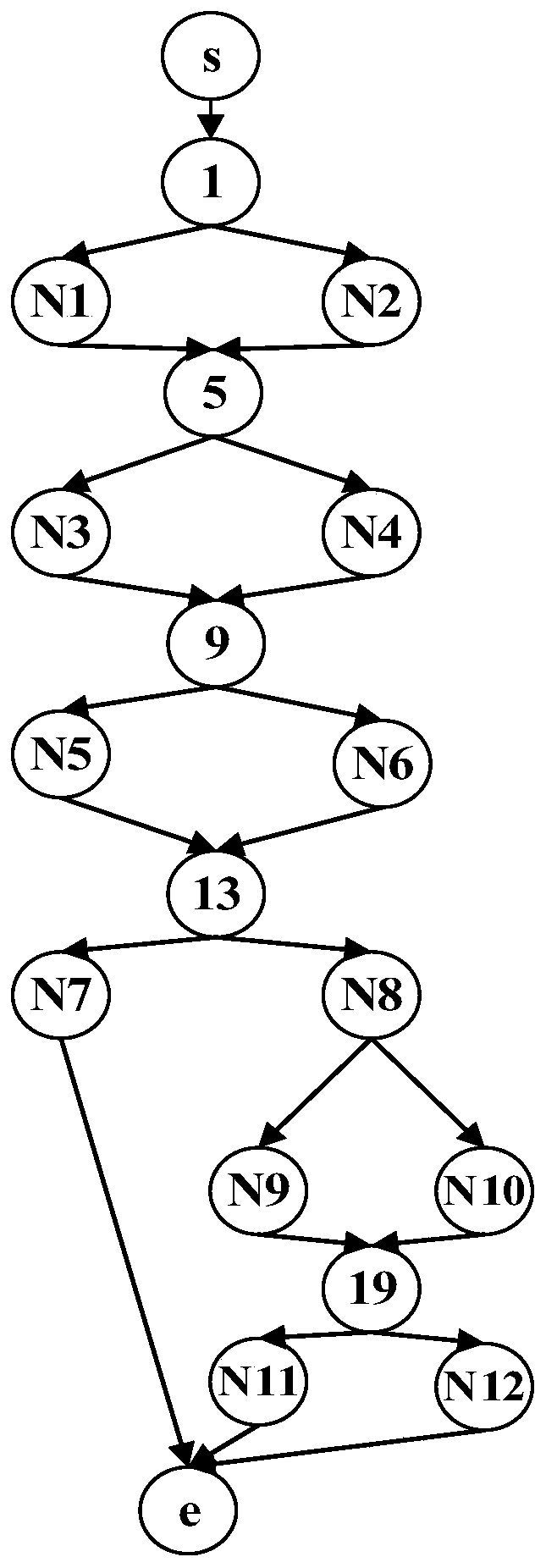
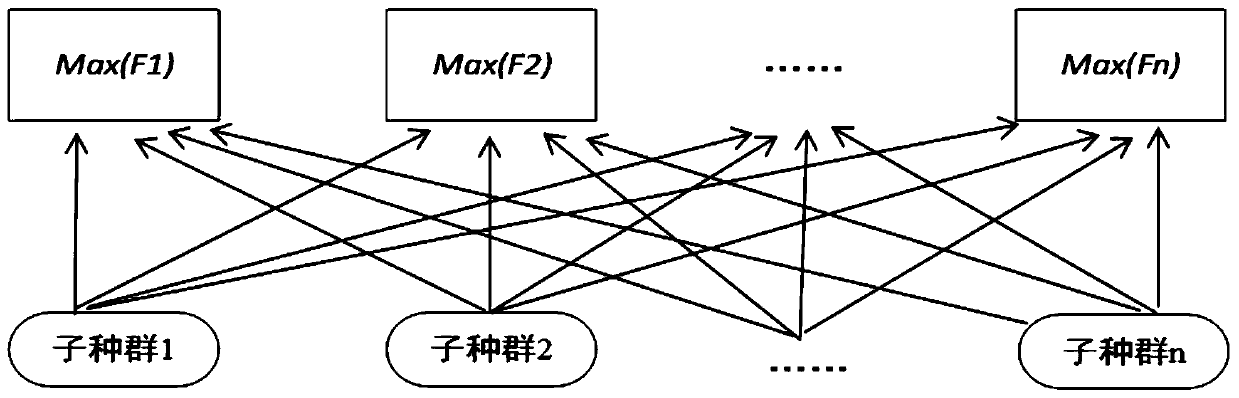
The invention relates to a software radio conformance test method based on a multi-population genetic algorithm, and solves the problem of loop test for some functions or the whole system. According to the method, according to the characteristics of the software radio conformance test, abstract component models are constructed, feasible test sequences are generated, a test sequence set capable ofrealizing test full coverage is constructed, and the test sequence set is optimized by adopting a multi-population genetic algorithm, so that the test efficiency is improved as much as possible, the test time is shortened, the vacancy of domestic software radio conformance test research is made up, and a good foundation is laid for subsequent test automation research.
Owner:NAT UNIV OF DEFENSE TECH
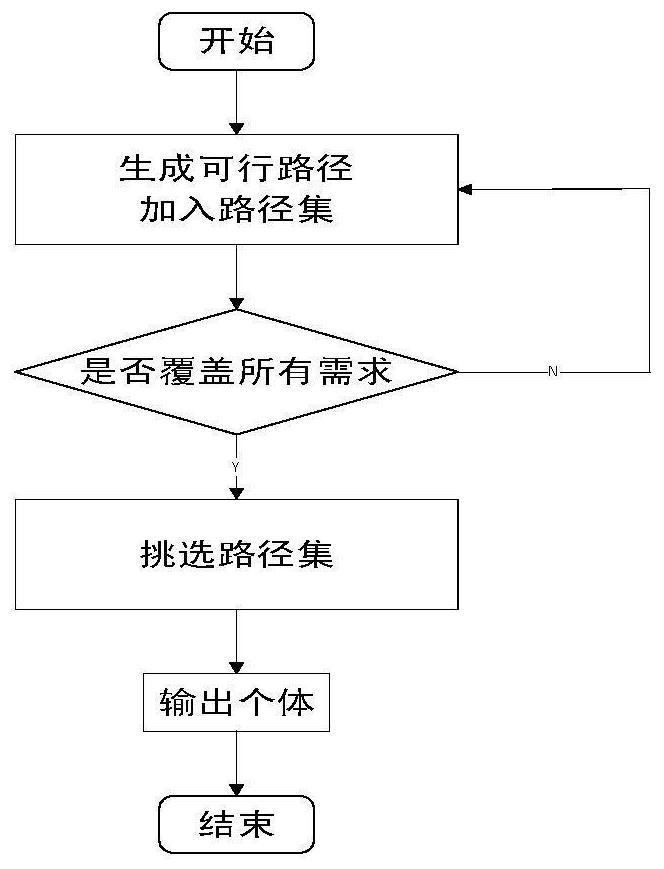
Multi-path coverage method and system combining key point probability and path similarity
ActiveCN111240995AImproved fitness functionImprove adaptabilityArtificial lifeSoftware testing/debuggingAlgorithmGenetics algorithms
The invention discloses a multi-path coverage method and system combining key point probability and path similarity. The method comprises the steps: firstly, dividing a theoretical path into an easily-covered path, a difficultly-covered path and an unreachable path; secondly, counting key point probabilities through the easy-to-cover path, calculating contribution degrees of individuals to generated test data according to the probabilities, improving a fitness function by utilizing the contribution degrees, and sorting target paths according to the key point probabilities; and finally, generating test data covering the target path by using a multi-population genetic algorithm, and continuously trying to cover the similar path of the target path after the sub-population covers the current target path in the evolution process. The fitness function is designed according to the probability of the key points, excellent individuals are protected, meanwhile, an individual information sharingstrategy is further perfected, individual resources in the population evolution process are reasonably utilized, it is avoided that too much time is wasted in the sub-population evolution process, andthus the test data evolution generation efficiency is effectively improved.
Owner:JIANGXI UNIVERSITY OF FINANCE AND ECONOMICS
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com