Method for identifying odontobulis mpotamophila family
A technology of sandfish and rivers, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., to reduce errors, avoid damage to fish bodies, and ensure accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
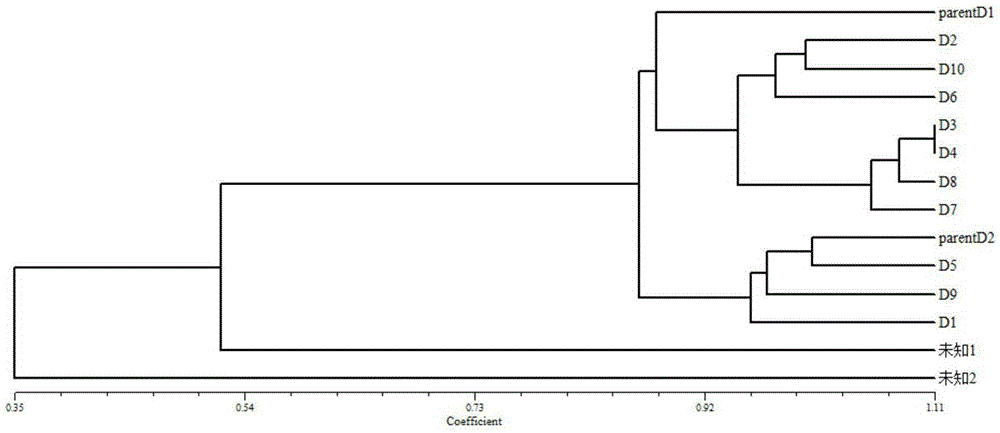
[0047] The 7 families to be tested are numbered as A, B, C, D, E, F, G, the parents and 10 offspring individuals of family D are randomly selected, and 2 individuals of river channa from unknown sources are selected, and recorded For unknown 1 and unknown 2, the genotype bp number of the individual to be tested is obtained according to the above method, and appropriate manual correction is performed to establish a family genotype information table. Table 4 shows the genotype of 12 pairs of primers among family D individuals data sheet.
[0048] Table 412 genotype data table for primers
[0049]
[0050]
[0051] Note: The number represents the size of the gene fragment
[0052] Convert the genotype data table into the POPGENE format required by the POPGENE1.32 software, use the POPGENE1.32 software to analyze the genetic distance of the individual to be tested, and obtain the genetic consistency, use EXCEL to transpose the genetic consistency table and convert it into ...
Embodiment 2
[0054] Randomly select the parents of 7 families and the offspring of the separated families, obtain the gene bp numbers of the parents and the offspring of each family according to the above method, and perform appropriate manual corrections to establish a family genotype information table, Table 5 It is the genotype data table of 12 pairs of primers among some individuals.
[0055] Table 512 genotype data table for primers
[0056]
[0057]
[0058] Note: The number represents the size of the gene fragment
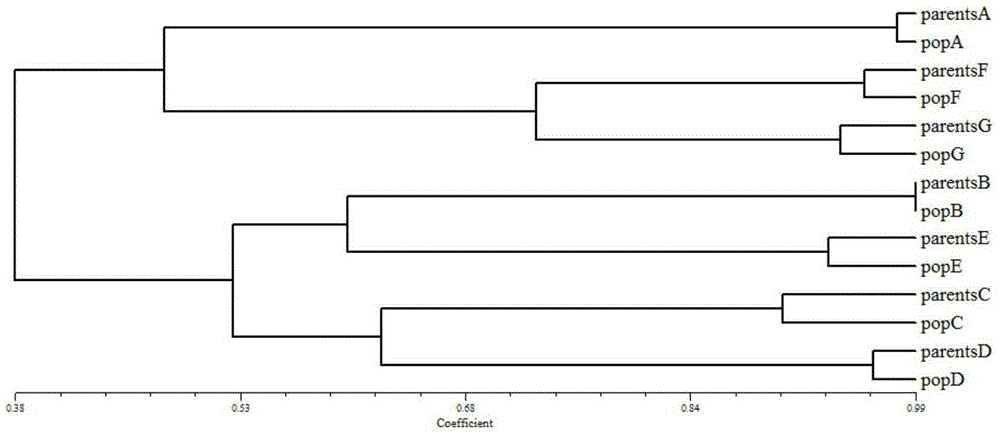
[0059] According to the method shown in Example 1, cluster analysis was carried out on the parents and offspring of 7 families, and a cluster diagram was obtained. figure 2 It is the clustering situation of 7 families. It can be seen from the clustering diagram that the parents and offspring groups of each family are correctly gathered together, which is completely consistent with the known situation. Therefore, the selected microsatellite markers can distinguis...
Embodiment 3
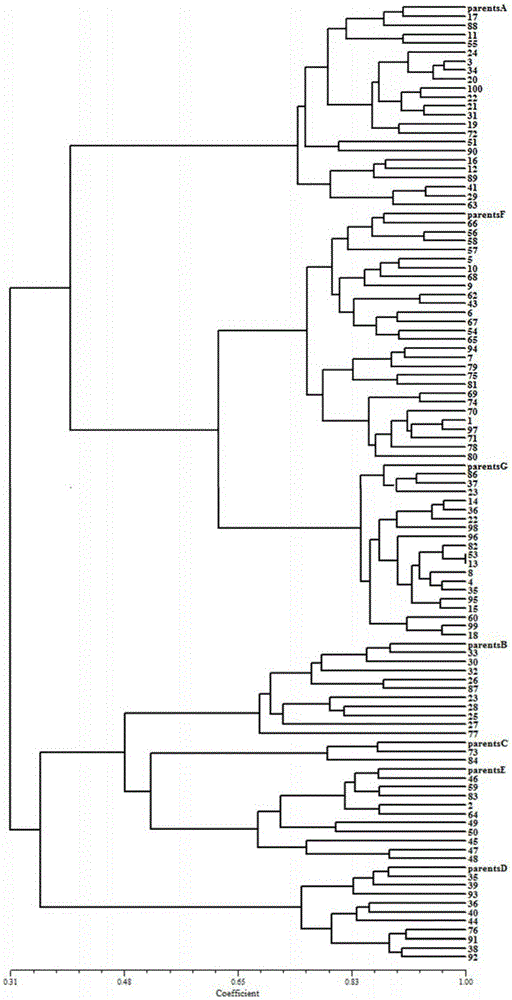
[0061] According to the method shown in Example 1, the genotype data of the parents of 7 families and 100 mixed offspring were obtained, and the genotype database was established, and the parents and offspring of the seven families were aggregated with POPGENE1.32, EXCEL and NTSYS software. class analysis, image 3 For the clustering case. It can be seen from the figure that all 100 mixed offspring can be assigned to the family of a certain parent, so it can be determined that all 100 mixed offspring individuals can find their corresponding parents.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com