Population genetic evolution map and construction method thereof
A population genetic and map technology, applied in the field of population genetic evolution map and its construction, can solve the problem of inaccurate population genetic evolution map
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
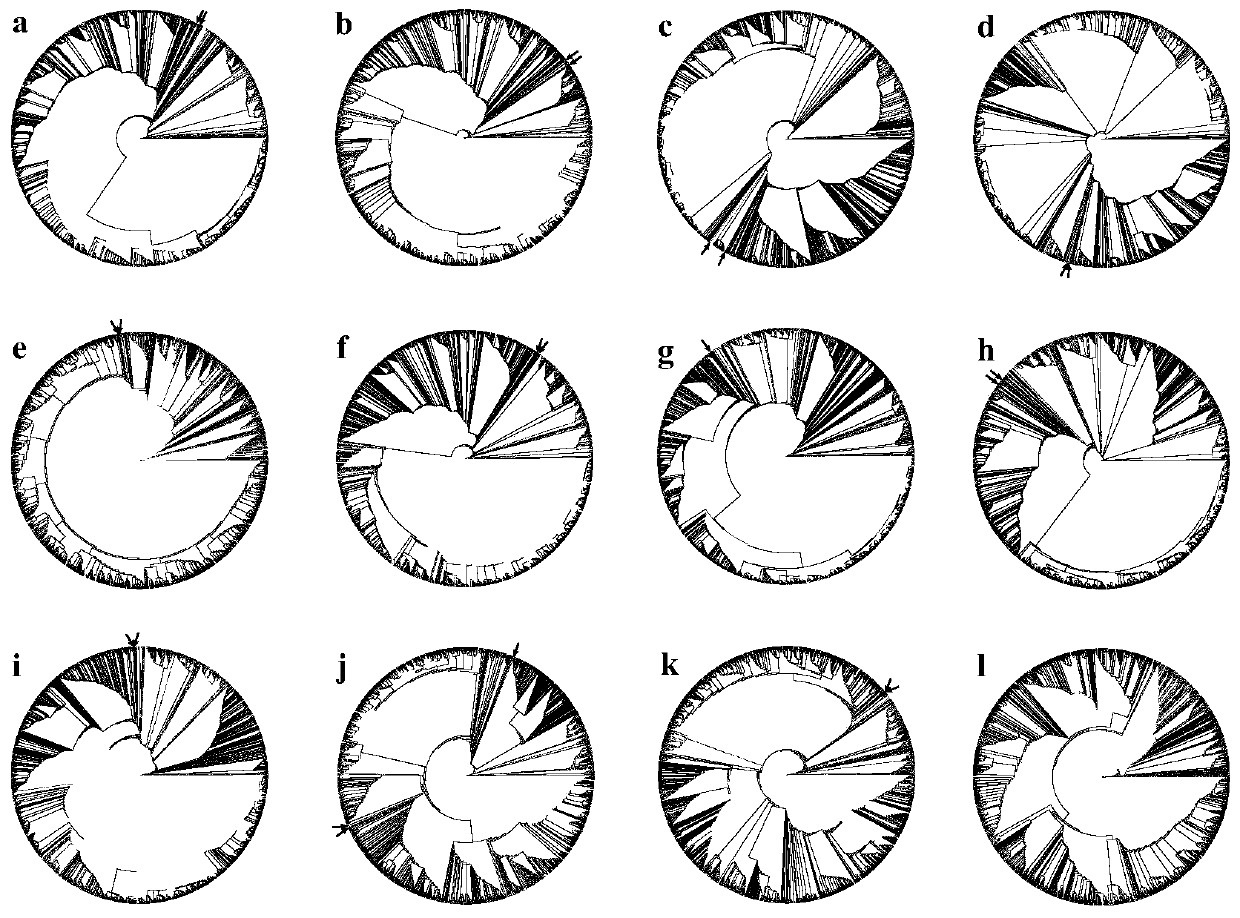
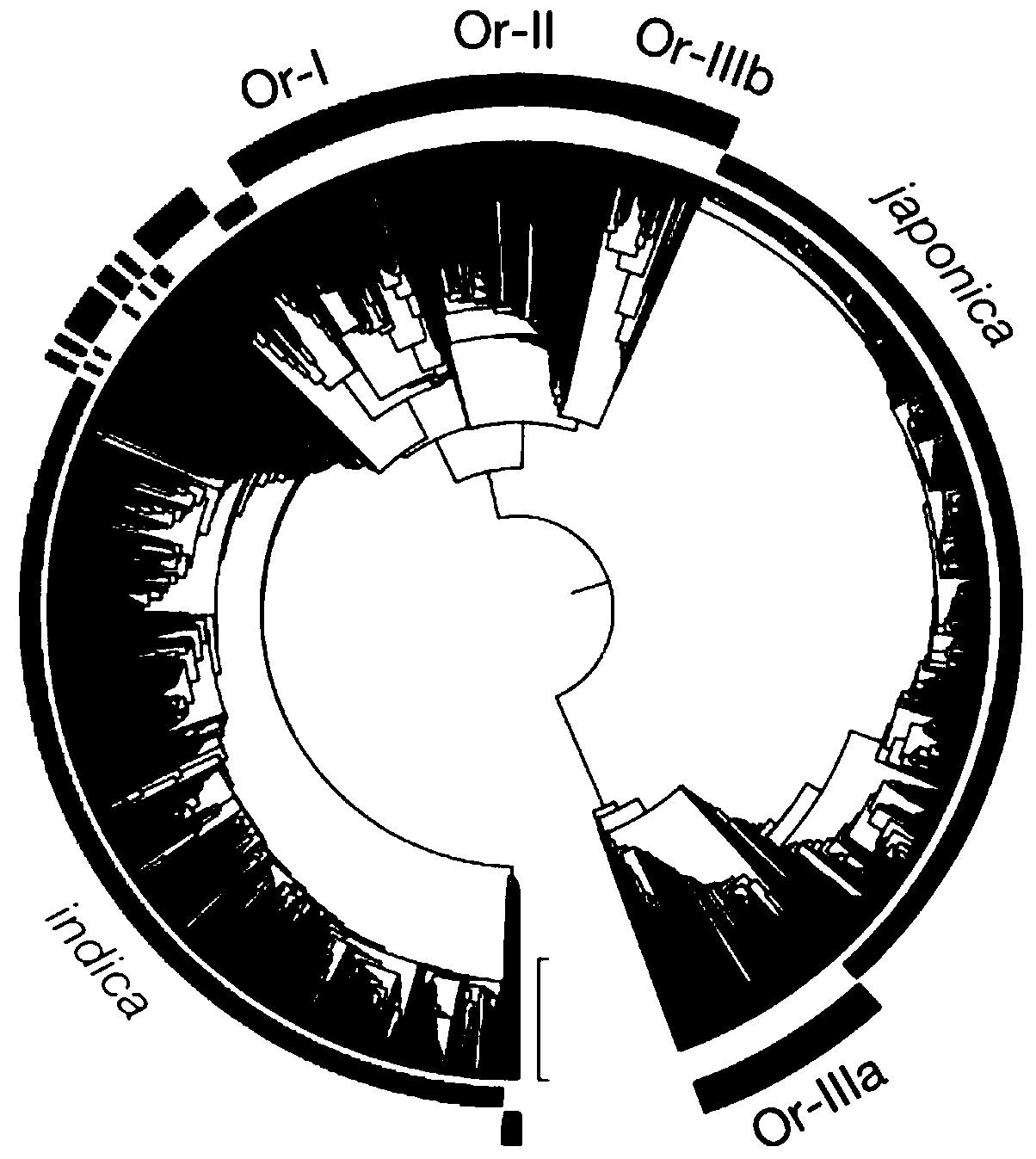
[0066] Example 1 Study on the evolution of rice using the region where recombination does not occur at the left and right ends of the rice centromere
[0067] (1) Determined the centromere position of the rice IRGSP 4.0 reference genome
[0068] The 1.155bp CentO satellite repeat sequence is the core element of rice centromere DNA. At the centromere of each chromosome in rice, CentO is variable in number, ranging in size from 65 kb to 2 Mb. Use the CentO sequence as the seed sequence to compare each chromosome of the IRGSP4.0 reference genome. According to the position information on the comparison, use the 10Kb window for quantitative statistics to determine the centromere boundary of each chromosome.
[0069] 2. The SNP distribution of the statistical data set, with a window of 10Kb.
[0070] 3. Select the SNP of the 200Kb sequence that does not recombine at both ends of the centromere.
[0071] The result is as figure 1 shown. figure 1 (See color picture reference pict...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com