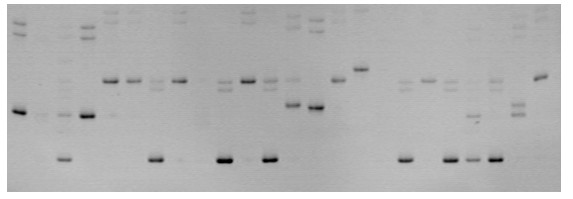
Detecting method for Litopenaeus vannamei Boone LvE165 microsatellite DNA marker
A detection method, the technology of Ive165, is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., to achieve the effect of simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] 1. Extraction of DNA from Litopenaeus vannamei
[0020] The materials of Litopenaeus vannamei used in this experiment were randomly extracted 24 DNA samples, and DNA was extracted from frozen muscle tissue. Cut 100mg of muscle tissue, mince it and place it in 0.7 ml of extraction buffer (6 M Urea (urea), 10 mM Tris-HCl, 125 mM NaCl (sodium chloride), 1% SDS (dodecyl muscle tissue) sodium sulfate), 10 mM EDTA (ethylenediaminetetraacetic acid), pH=7.5), add proteinase K (20 mg / ml) at a final concentration of 0.1 mg / ml, and digest overnight at 37°C. The reactant was extracted three times with phenol:chloroform (1:1), and the extracted supernatant was extracted once with an equal volume of chloroform. The supernatant was precipitated with isopropanol, and then dissolved in 500 1μl×TE (10 mM Tris.HCl, 1 mM EDTA, pH=8.0). Treat the DNA sample with RNase (20 μg / m1) at 37°C for 1 hour, and then use phenol / chloroform extraction solution for DNA purification. Extract once with ...
Embodiment 2
[0027] Screening of Microsatellite Loci and Determination of Polymorphic Markers
[0028] 1. Source of microsatellite loci and screening of microsatellite sequences
[0029]Collect and download (in FASTA format) existing EST sequences from the NCBI (http: / / www.ncbi.nlm.nih.gov) database, use SSRhunter software to search and analyze the 5,736 sequences obtained one by one, for 2-6 bases Microsatellite DNA sequences with basic repeating units and repeats greater than 5 were isolated. The ESTs containing microsatellites were clustered and analyzed using SeqMan in the software DNAstar, and the ESTs clustered in different contigs were selected to design primers.
[0030] 2. Design of Microsatellite Marker Primers
[0031] Use primer design software Primer Premier 5.0 and Primerselect in DNAstar to design primers on the flanking sequences of microsatellite repeats; primers are required to meet the following conditions: (1) The length of the primer is 17-25bp; (2) The GC content is...
Embodiment 3
[0040] 1. DNA extraction of Litopenaeus vannamei
[0041] The 10 Litopenaeus vannamei used in this example were extracted from muscle tissue soaked in 95% ethanol. Wash twice with ddH2O before extraction, and then place in 0.7 ml of extraction buffer (6 M Urea (urea), 10 mM Tris-HCl. 125 mM NaCI (sodium chloride), 1% SDS (dodecyl muscle Sodium sulfate), 10 mM EDTA (ethylenediaminetetraacetic acid), pH=7.5), add proteinase K (20 mg / ml) at a final concentration of 0.1 mg / ml, cut into pieces, and digest overnight at 37°C. The reactant was extracted three times with phenol:chloroform (1:1), and the extracted supernatant was extracted once with an equal volume of chloroform. The supernatant was precipitated with isopropanol, and then dissolved in 500 1μl×TE (10 mM Tris.HCl, 1 mM EDTA, pH=8.0). Treat the DNA sample with RNase (20 μg / m1) at 37°C for 1 hour, and then use phenol / chloroform extraction solution for DNA purification. Extract once with phenol / chloroform and once with chl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com