Patents
Literature
37results about How to "Accurate amplification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Primer combination and kit for simultaneous detection of 93 bovine genetic defect genes and lethal haplotypes
ActiveCN107099607AAccurate amplificationImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceAssortative mating
The invention discloses a primer combination and a kit for simultaneous detection of 93 bovine genetic defect genes and lethal haplotypes. Through use of the primer combination and the kit, casual mutation sites (SNP and insertion or deletion of short fragments) of the 93 genetic defect genes (such as PIRM syndrome, crooked tail syndrome, epidermolysis bullosa and the like) and the lethal haplotypes (HH1, HH3, HH4, HH5 and JH1) can be detected in one time, cattle with monogenic inheritance defect gene carriers and lethal haplotype carriers can be screened, and accurate guidance is provided for cattle breeding and genetic evaluation, herd selection and assortative mating, population genetic improvement, cultivation of new varieties and genetic resource protection. The primer combination and the kit have the characteristics of high throughput, low cost, high accuracy, and the like, is suitable for the varieties of beef cattle and dairy cattle, and can be widely used in the breeding and reproduction field of the cattle.
Owner:DAIRY CATTLE RES CENT SHANDONG ACADEMY OF AGRI SCI
Primer for amplifying COI sequence of DNA code bar of viper species, PCR method and kit
InactiveCN104164426AEfficient amplificationAccurate amplificationMicrobiological testing/measurementFermentationMolecular identificationElectrophoresis
The invention belongs to the technical field of viper species and traditional Chinese herbal identification, and discloses a primer for amplifying the COI sequence of DNA code bar of viper species, a PCR method, and a kit; wherein the method comprises the steps of DNA extraction, PCR amplification, and electrophoresis detection. The identification method has the characteristics of convenience, little DNA using amount, and good universality; and overcomes the shortages of COI sequence universal primers (LCO1490 and HCO2198) in viper gene amplification and traditional Chinese herbal identification in the prior art. The primer can be applied to the amplification of COI sequence of viper, and establishes a foundation for molecular identification of viper.
Owner:晁志
Scophthalmus maximus T170G single nucleotide polymorphic marking detection method
InactiveCN102586454AAccurate amplificationGood polymorphismMicrobiological testing/measurementNucleotideGenotype
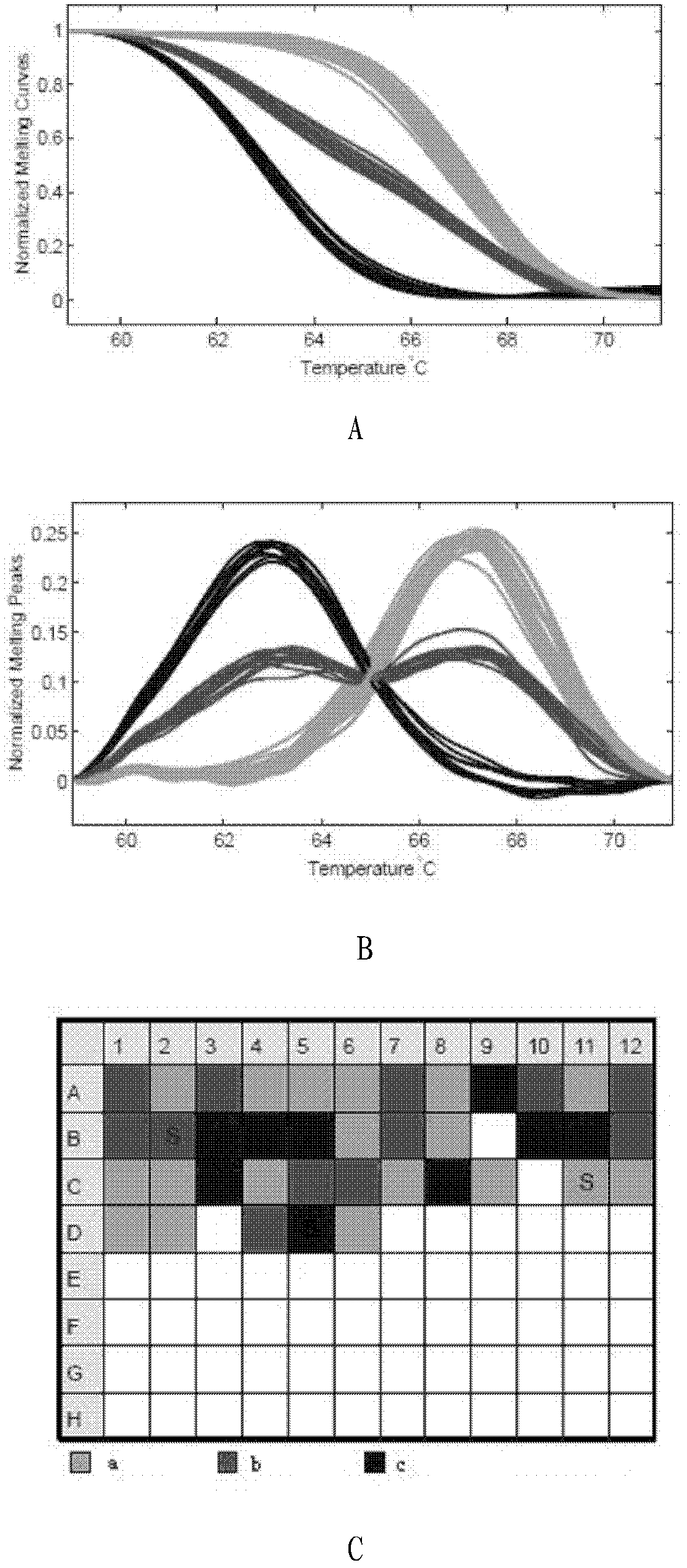
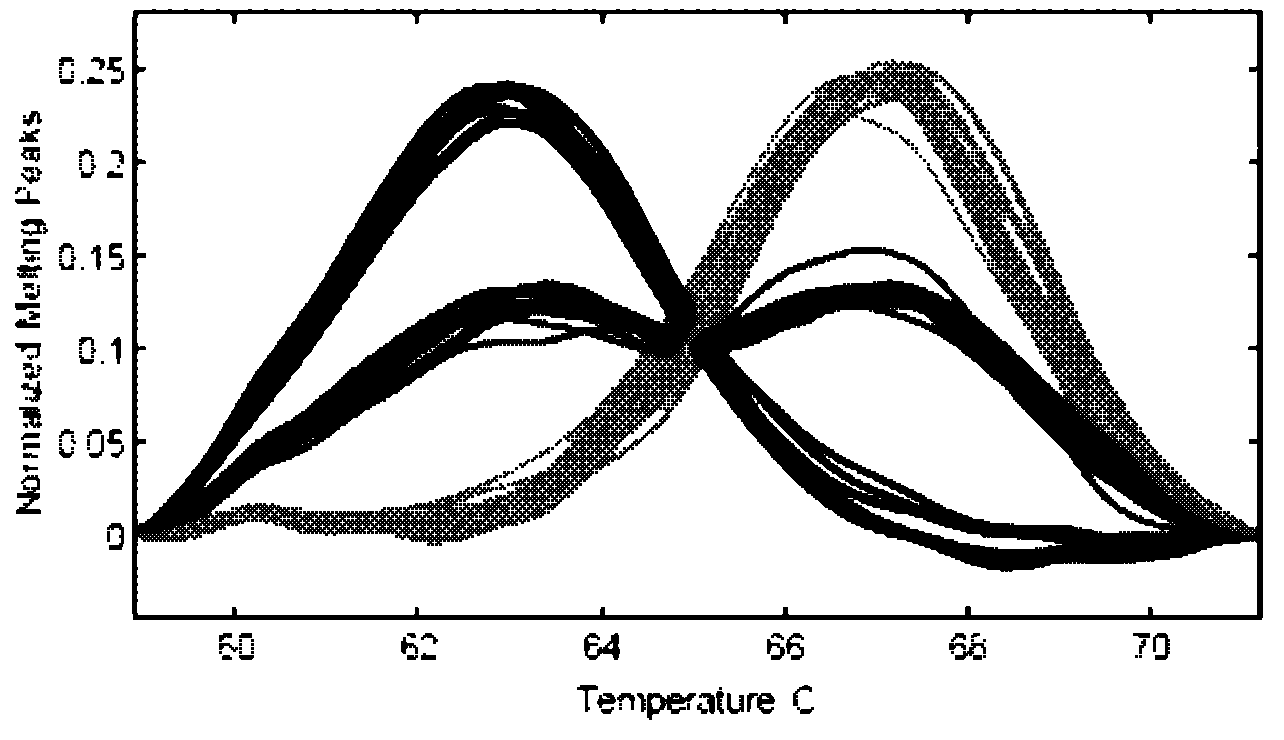
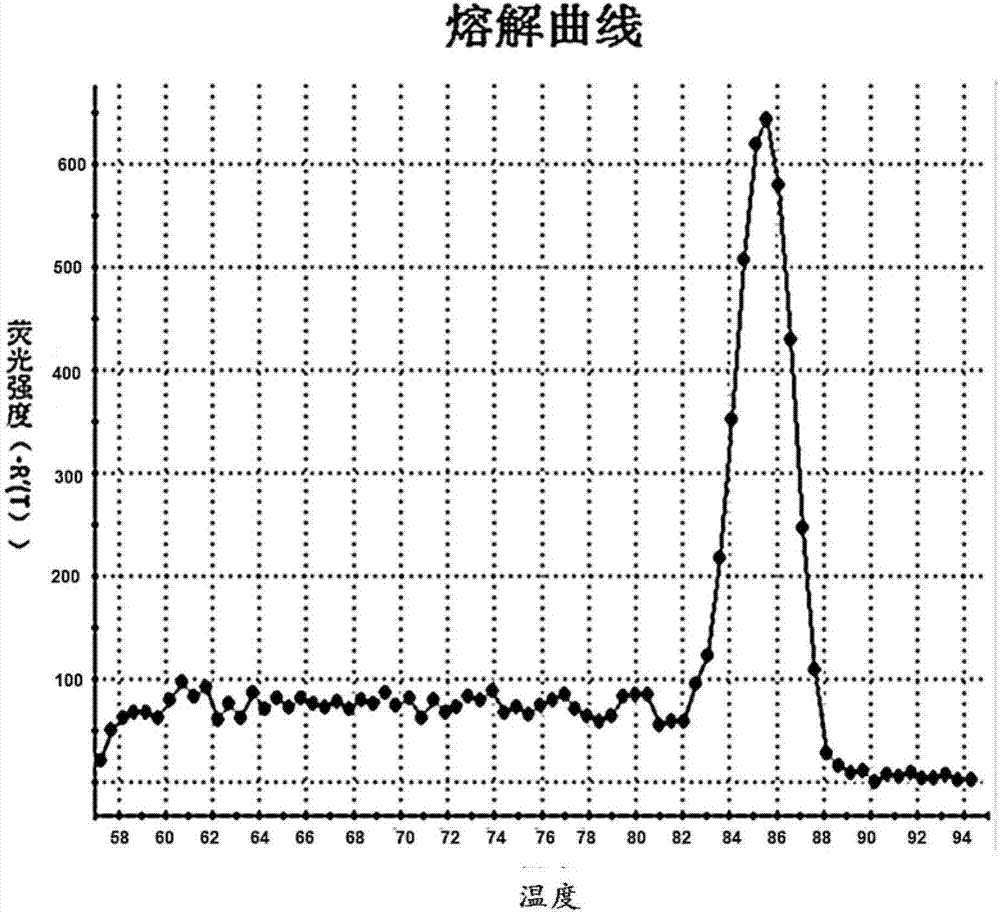
The invention relates to a scophthalmus maximus T170G single nucleotide polymorphic marking detection method. The method comprises the following steps: extracting scophthalmus maximus genome DNA and diluting for later use; analyzing the sequences of EST, screening the sequence containing a candidate SNP locus, designing a specific primer at the two ends and designing a non-marking probe with the closed 3' end in front of and behind the locus (comprising the locus); performing asymmetric PCR amplification on the scophthalmus maximus group genome DNA by using the primer; hybridizing the amplification product with the non-marking probe with the closed 3' end; and placing the hybrid product on a LightScanner, and detecting and analyzing the melting curve to obtain the genetic polymorphism mapof the scophthalmus maximus. The method is applicable to detection technologies of scophthalmus maximus genetic marking, genealogy authentication, genetic linkage map construction and the like. According to the method which is convenient, quick and accurate, the scophthalmus maximus T170GSNP marked genetic variation map can be obtained quickly; and the genotype of each individual of the scophthalmus maximus can be detected intuitively.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Poppy species specificity genetic marker detecting system
ActiveCN105861711AAccurate amplificationRapid amplificationMicrobiological testing/measurementDNA/RNA fragmentationStatistical analysisBiology
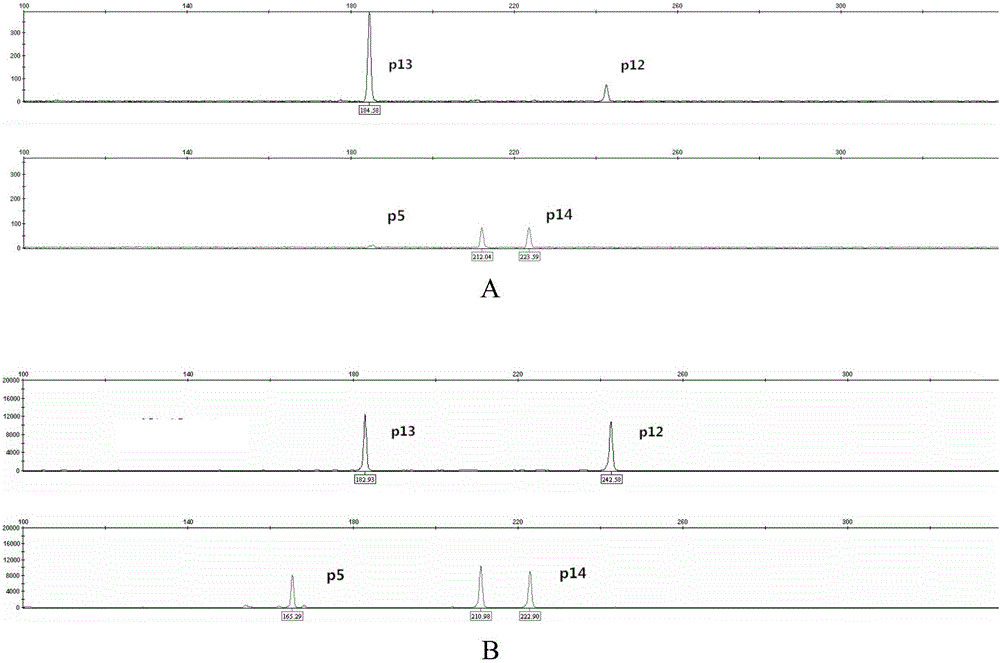
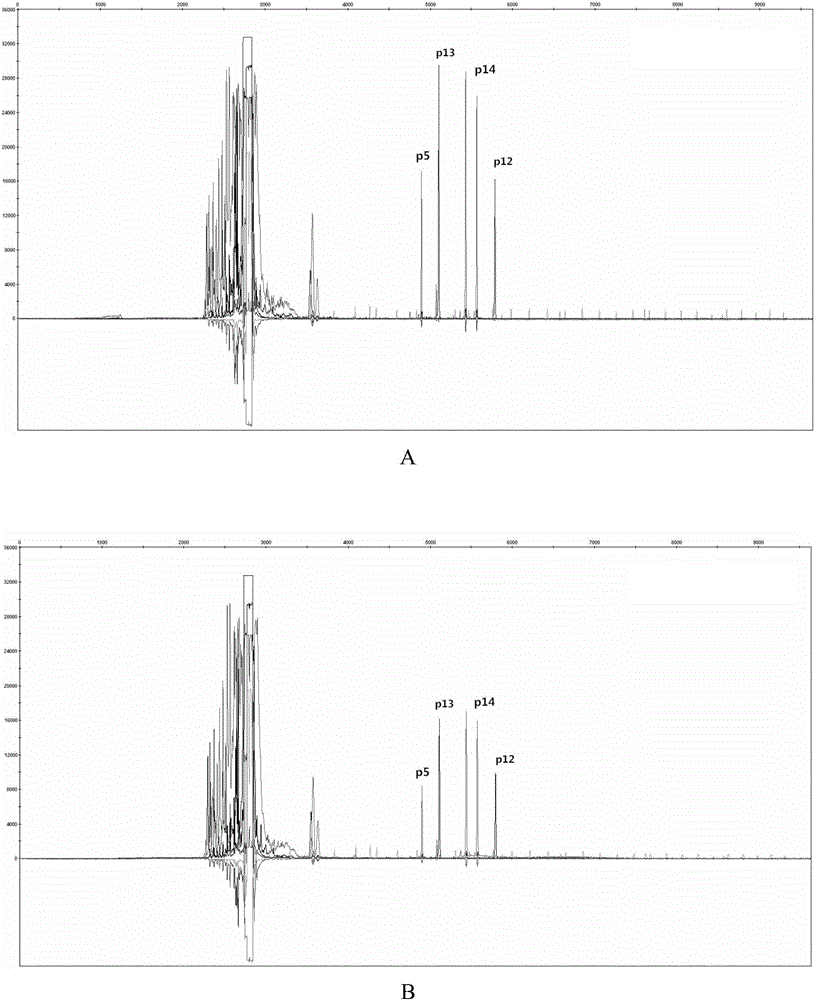
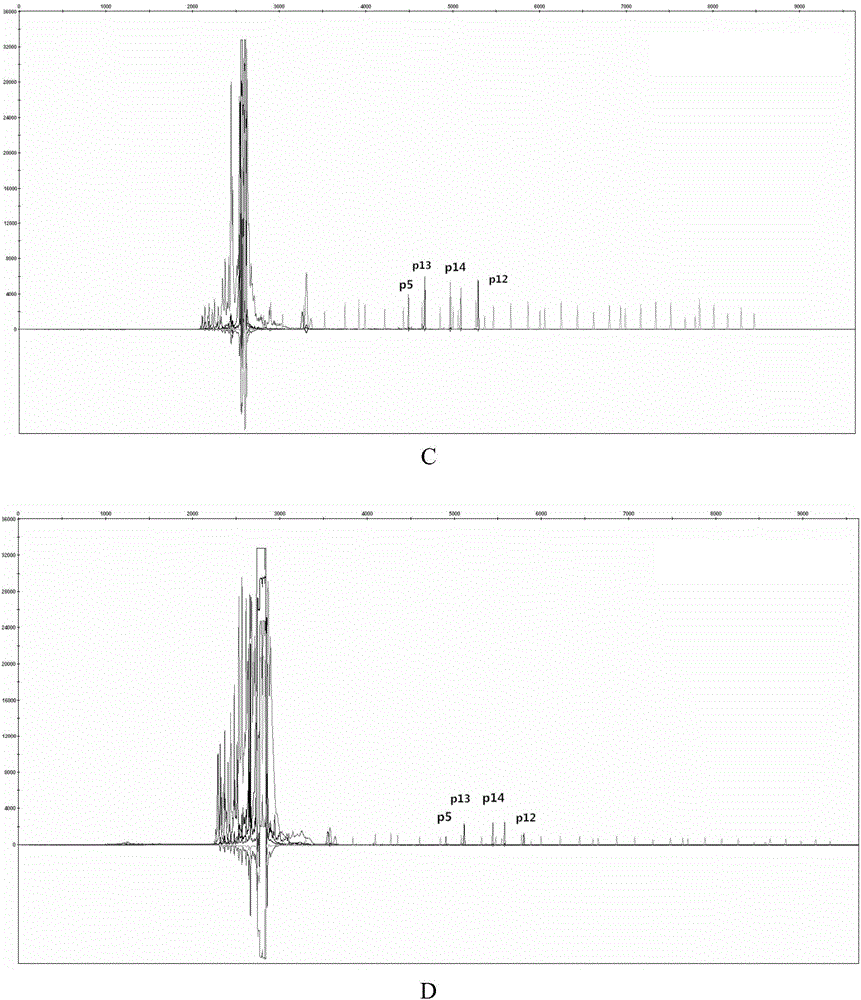
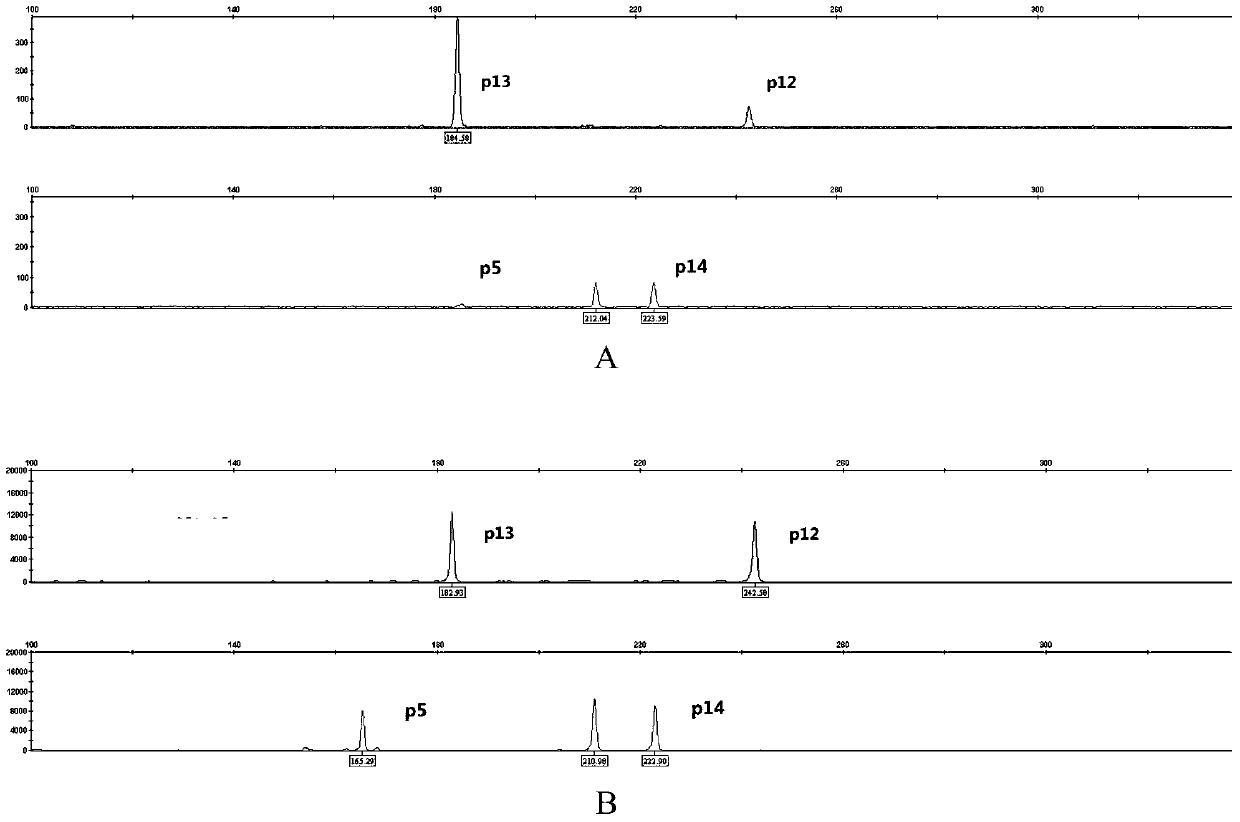
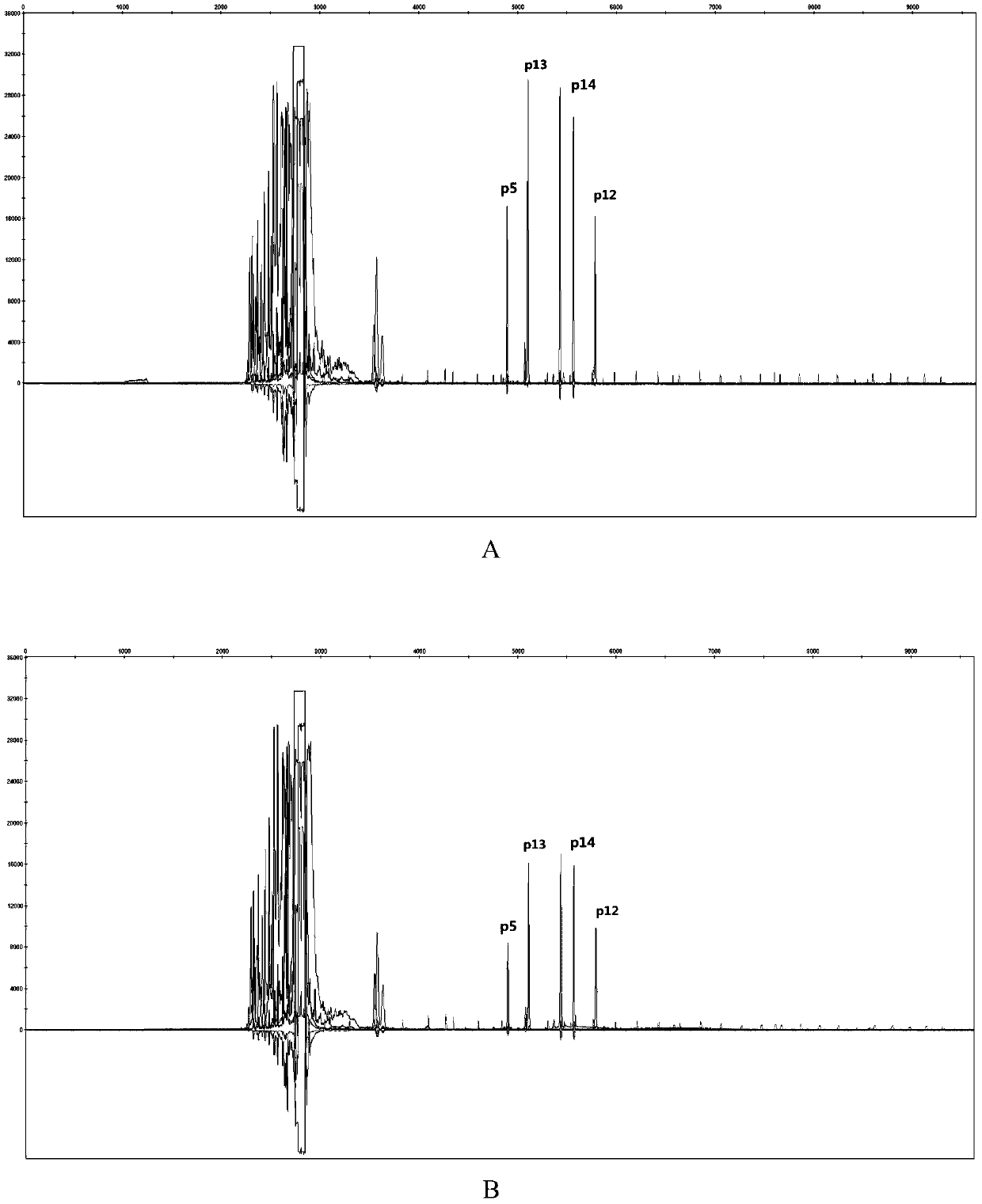
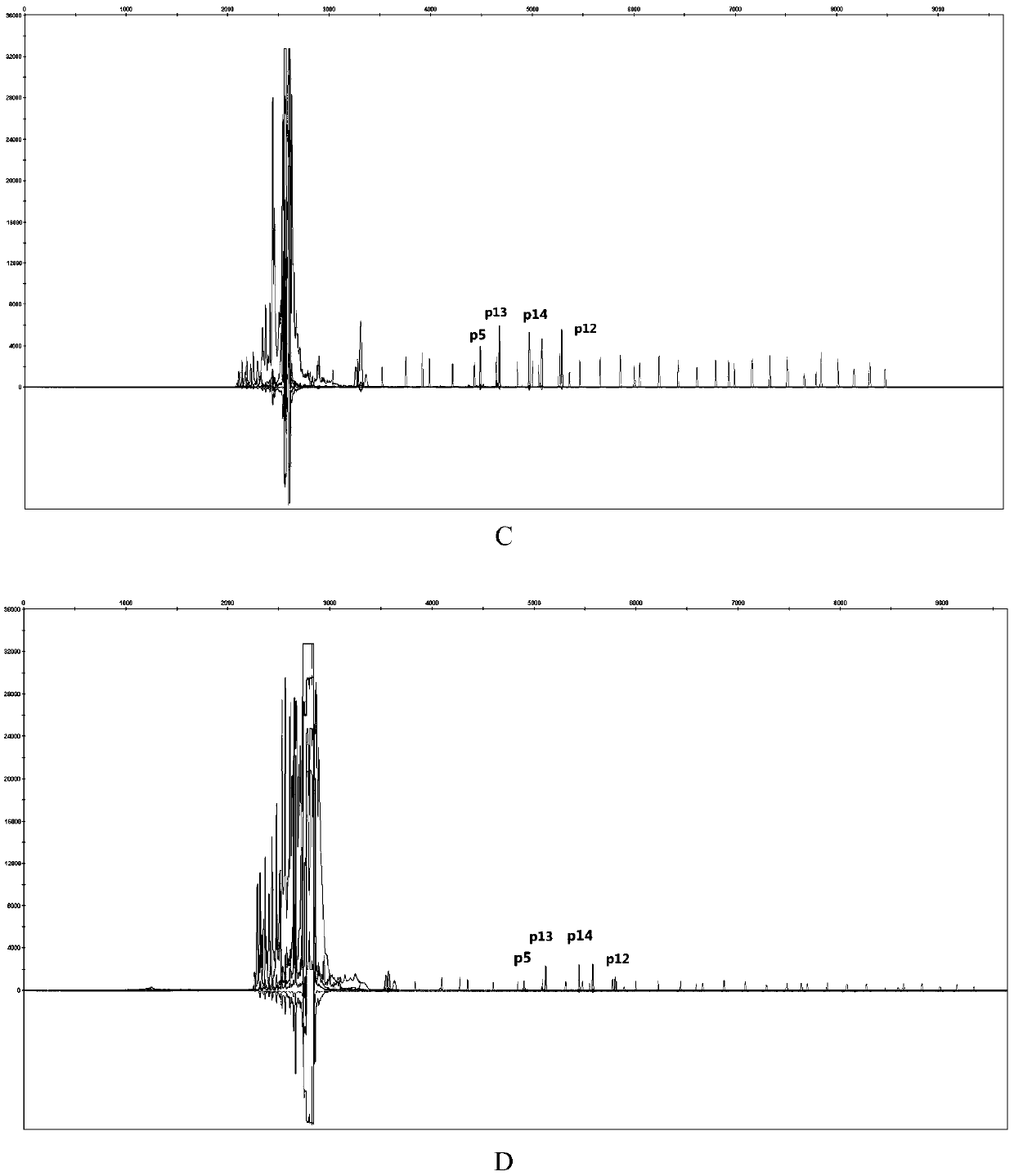
The invention discloses a poppy species specificity genetic marker detecting system. A primer pair set for identifying poppy is provided and is composed of a primer pair P12 (sequence 1 and 2), a primer pair P13 (sequence 3 and 4), a primer pair P5 (sequence 5 and 6) and a primer pair P14 (sequence 7 and 8). Four STR loci of poppy are amplified at the same time, and the amplification is more accurate and rapid compared with amplification of a single locus; the system has higher species specificity when identifying poppy and affinis plants, and an effective method is provided for further studying poppy STR loci and inspecting and identifying the poppy species in a case; in an optimized poppy STR composite amplification system, amplification of all loci can be balanced, sensitivity is high, stability is high, and frequently-seen corrosion check materials in a drug-related case can be easily inspected; accuracy is high, parting of four loci of each of poppy samples from different producing areas can be achieved through detection and data statistic analysis.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Method for detecting nucleic acid using asymmetric isothermal amplification of nucleic acid and signal probe
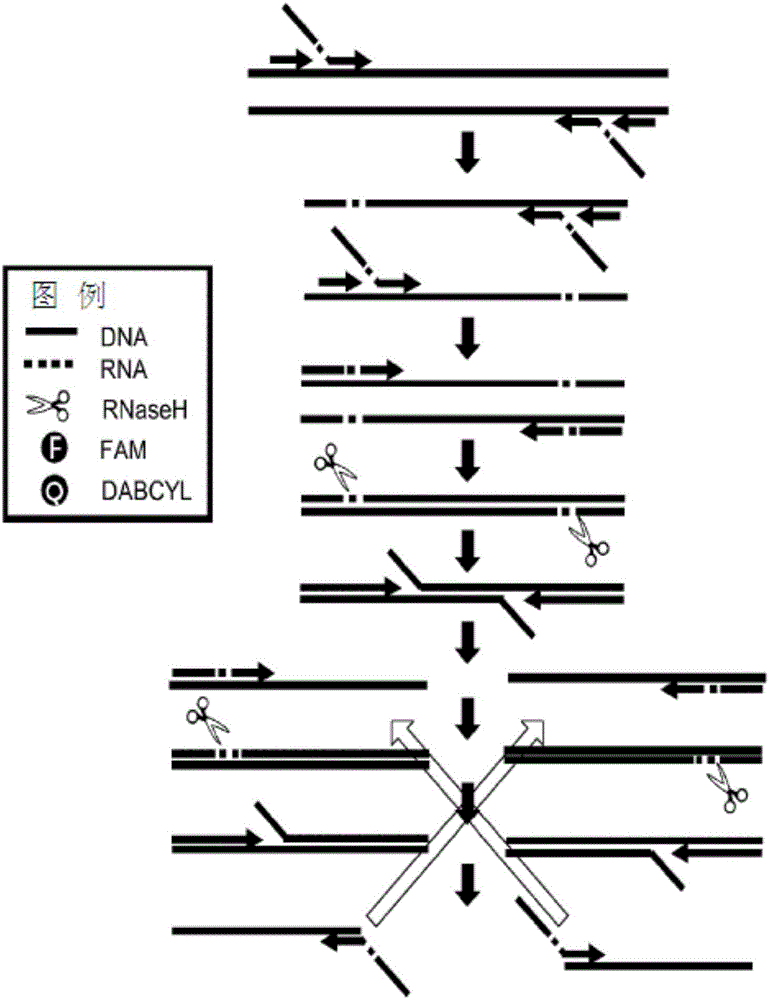
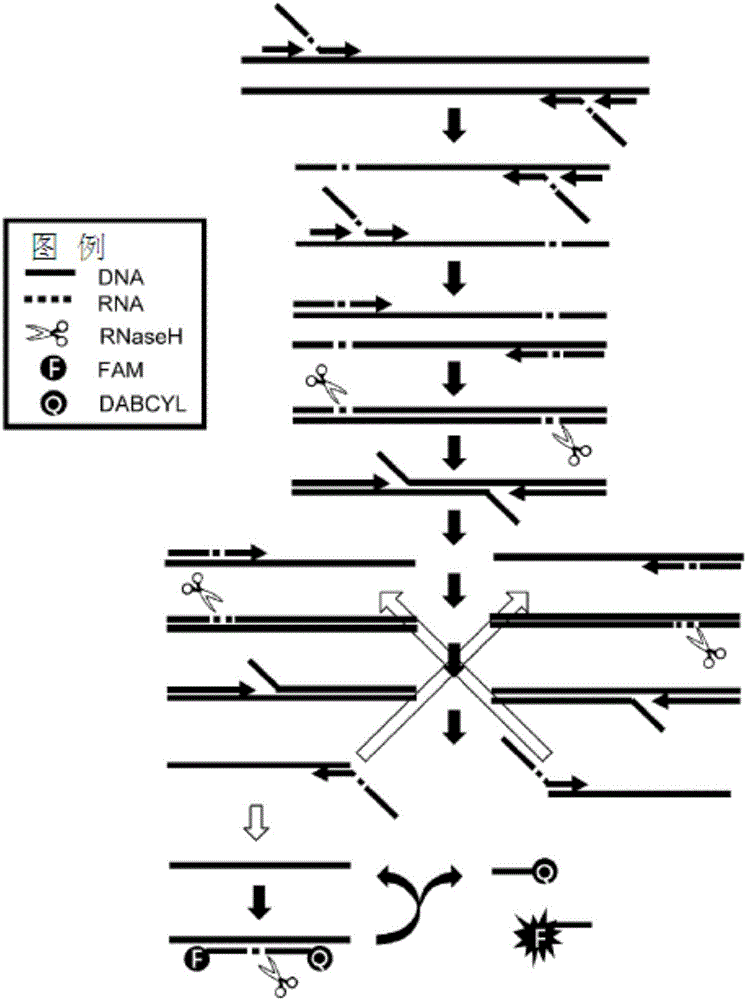
InactiveCN105960467ARapid and Accurate AmplificationAccurate amplificationMicrobiological testing/measurementDiseaseA-DNA
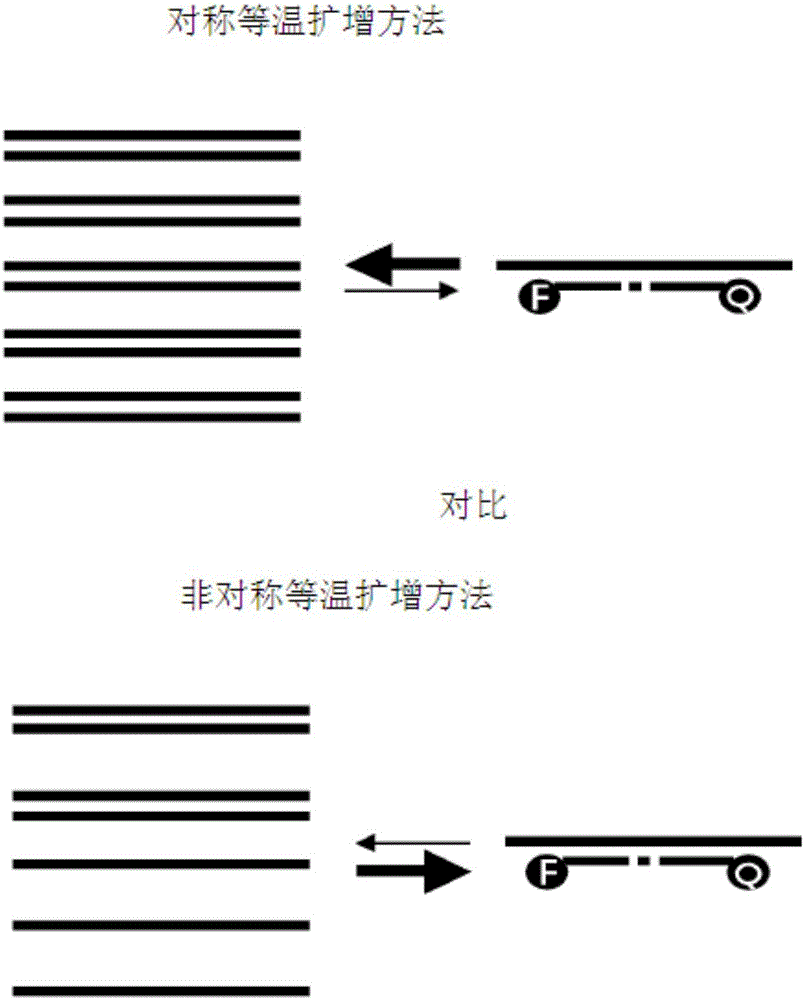
The present invention relates to a method for accurately detecting a target nucleic acid by asymmetrically and isothermally amplifying the target nucleic acid using an external primer set, an internal primer set having different percentages of forward and reverse DNA-RAN-DNA hybrid primers, and a DNA-RNA-DNA hybrid signal probe and amplifying a signal of the probe at the same time. According to the present invention, the signal of the probe can be efficiently amplified compared with the method of the prior art, a symmetric iTPA method, which is an isothermal primer and probe amplification method using the same percentage of primers. Therefore, the present invention is applied to the accurate detection and confirmation of pathogens, detection of gene modification inducing identified phenotypes, diagnosis of susceptibility to genetic diseases, evaluation of gene expression, and various genome projects, and thus is useful in the molecular biological researches and disease diagnosis.
Owner:DXGENE
Soybean branch number molecular marker and application thereof
InactiveCN111334602AAccurate locationAccurate amplificationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyBase sequence
The invention discloses a soybean molecular marker, a method for obtaining the soybean molecular marker and an application of the soybean molecular marker. The molecular marker is obtained by adoptinga map-based cloning mode; genome DNA is amplified by using a specific primer pair; and the obtained molecular marker is applied to identification of soybean branch number. The map-based cloning method is utilized to locate and clone a multi-branch decisive base sequence in a multi-branch soybean genome, and whether soybeans are multi-branch soybeans or not is determined according to a cloning result, so the soybeans are planted, and the yield of the soybeans is increased.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Primers and kit for detecting macrolide drug-resistance genes of bacteria
ActiveCN104561341ARapid expansionAccurate amplificationMicrobiological testing/measurementDNA/RNA fragmentationBacteroidesMicroorganism
The invention discloses primers and a kit for detecting the macrolide drug-resistance genes of bacteria, and belongs to the technical field of microbiological detection. The sequences of the primers for detecting the macrolide drug-resistance genes of bacteria provided by the invention are shown in SEQ ID NO: 1-6, and the three primer pairs are capable of specifically amplifying the three macrolide drug-resistance genes of ermA, ermB and ermC of multiple bacteria. The invention further discloses a kit for detecting the macrolide drug-resistance genes of bacteria, wherein the kit contains an EvaGreen fluorescent dye, and is capable of rapidly and accurately detecting the macrolide antibiotic-related drug-resistance genes of bacteria. The primers and the kit disclosed by the invention have the advantages of simplicity and convenience in operation, high specificity, high sensibility, low cost, high flux and the like, and can be used for rapidly detecting macrolide antibiotic-related drug-resistance genes of clinic pathological bacteria and providing a reference for clinic treatment.
Owner:CHONGQING DIAN SRAB CENT FOR CLINICAL LAB CO LTD
Method for detecting C135T single nucleotide polymorphism of turbot
InactiveCN102534043AAccurate amplificationGood polymorphismMicrobiological testing/measurementExpressed sequence tagGenetic linkage map
The invention discloses a method for detecting C135T single nucleotide polymorphism (SNP) of turbot. The method comprises the following steps of: extracting genome DNA of turbot and diluting for later use; analyzing a sequence of an expressed sequence tag (EST), screening a sequence containing a candidate SNP locus, designing specific primers at two ends of the sequence, and designing 3' end-closed unlabeled probes before and after the locus (including the locus); performing asymmetrical polymerase chain reaction (PCR) amplification on population genome DNA of turbot by using the primers; hybridizing an amplification product and the 3' end-closed unlabeled probes; and detecting a melting curve of the hybrid on a light scanner, and analyzing the melting curve to obtain a genetic polymorphism map of turbot. The method is suitable for detection technologies such as turbot population genetic markers, lineage authentication, genetic linkage map construction and the like; and by the method,a genetic variation map of the C135TSNP of turbot can be quickly, easily and accurately obtained, and the genotype of each individual of turbot can be intuitively detected.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Primer set and application thereof in amplifying SIV/SHIV (simian immunodeficiency virus/simian human immunodeficiency virus) genome, and kit
ActiveCN105624153AGood sensitivityIncreased sensitivityMicrobiological testing/measurementDNA preparationNon specificSimian human immunodeficiency virus
The invention relates to the field of molecular biology, particularly a primer set and application thereof in amplifying SIV / SHIV (simian immunodeficiency virus / simian human immunodeficiency virus) genome, and a kit. The primer set provided by the invention can be used for specific, stable and accurate amplification to obtain the target sequence. The agarose gel electrophoresis detection indicates that no non-specific strip is generated, and no trailing or dispersion is generated. Compared with the SIVmac239 whole genome sequence registered in Genbank after sequencing, the matching degree is 100%, and no base replacement or deletion (gap) is detected. The experiment indicates that the primer set has favorable sensitivity for the SIV / SHIV genome and can have favorable detection effects when the virus copy number is 100.5-1.0, and thus, the analysis judges that the sensitivity difference is related to virus sequence variation degree.
Owner:INST OF LAB ANIMAL SCI CHINESE ACAD OF MEDICAL SCI +1
Specific primer for quantitatively detecting yellow leaf curl virus, as well as application of specific primer
InactiveCN107058626AQuantitatively accurateAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationTomato yellow leaf curl virusPathogenic bacteria
The invention belongs to the field of pathogen detection, and provides a primer pair for quantitatively detecting a yellow leaf curl virus, application of the primer pair to quantitative detection of the yellow leaf curl virus and a method for quantitatively detecting the yellow leaf curl virus. The primer pair for quantitatively detecting the yellow leaf curl virus can be used for efficiently and accurately amplifying a target fragment. The method for quantitatively detecting the yellow leaf curl virus by virtue of the primer pair can be used for accurately and rapidly identifying pathogenic bacteria and accurately quantifying a viable count.
Owner:BEIJING PLANT PROTECTION STATION
Yanbian yellow cattle meat quality-related ANGPT4 gene SNP molecular marker, primer pair, kit and application thereof
ActiveCN110541036AAccurate amplificationFood processingMicrobiological testing/measurementNucleotideCorrelation analysis
The invention provides a Yanbian yellow cattle meat quality-related ANGPT4 gene SNP molecular marker, a primer pair, a kit and an application thereof, and belongs to the technical field of cattle meatquality screening. The Yanbian yellow cattle meat quality-related ANGPT4 gene SNP molecular marker contains a nucleotide sequence in which the polymorphism of 41995bp of a gene is C / T. The polymorphism of the ANGPT4 gene and the meat quality trait are subjected to correlation analysis; the 41995bp C / T site has the significant correlation with the characters of the fat content, the protein content, the carcass weight and the marble pattern grade of Yanbian yellow cattle; and it is showed that the fat content, the carcass weight and the marble pattern grade of a CT genotype are obviously higherthan those of a CC genotype. The protein content of the CC genotype is significantly higher than that of the CT genotype. The SNP molecular marker of the ANGPT4 gene can be used as a meat quality trait screening-related gene marker and is used for high-grade beef cattle early-stage selection of the Yanbian yellow cattle.
Owner:YANBIAN UNIV
DNA (deoxyribonucleic acid) bar code and application of bar code to identification of muscadine
ActiveCN107779521AVersatileImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationAgricultural science
The invention discloses a DNA (deoxyribonucleic acid) bar code. The nucleotide sequence of the DNA bar code is as shown in SEQ ID NO.1. According to the DNA bar code, an ITS sequence of a muscadine isfirstly disclosed, available molecular markers are provided for species identification of the muscadine, and the DNA bar code further comprises parts of conserved sequences and has universality whenbeing used for species identification of grapes. The invention discloses a method and a primer for preparing the nucleotide sequence of the DNA bar code and can acquire sequences of an ITS area of themuscadine. The invention further discloses a molecular identification method of the muscadine and a detection primer and a kit for identifying the muscadine. PCR (polymerase chain reaction) amplification is performed by the aid of the detection primer or the kit to obtain sequences of ITS areas of the grapes, an amplification sequence and the sequence of the DNA bar code of the muscadine are compared or applied to construction of phylogenetic trees of the grapes, so that grape varieties are accurately identified, and identifying processes are simple to operate, high in efficiency and good inrepeatability.
Owner:NANJING XIAOZHUANG UNIV
Universal method for detecting copy number of target gene and titer of virus and application
PendingCN112176100AEnsure accurate amplificationGuaranteed accuracyMicrobiological testing/measurementDNA/RNA fragmentationTiterGenetic testing
The invention belongs to the technical field of biology, and particularly relates to a universal method for detecting the copy number of a target gene, a universal method for detecting the titer of alentivirus, application of the universal method, a PCBP2 gene detection kit and a universal kit for detecting the copy number of the target gene and the titer of the lentivirus. A PCBP2 gene is adopted as a calibration factor, and the nucleotide sequence of a PCBP2 gene template is shown as SEQ ID: NO 1 or SEQ ID: NO 2. The PCBP2 gene solves the problems that a traditional method can only detect ahuman sample and has universality; the traditional method for detecting the titer of a virus is poor in coincidence with a method for detecting the titer of the virus through a flow cytometry, and the universality of detecting the titer of the virus in practical clinical application is not high; and the PCBP2 gene is adopted as the calibration gene, the result stability is good, detection of thetiter of the lentivirus is more accurate, the detection result of the lentivirus is more coincident with the detection result of the flow cytometry, and clinical application is facilitated.
Owner:CHONGQING PRECISION BIOTECH CO LTD +1
Primer group capable of accurately measuring ITS base sequence of cerasus schneideriana, synthesis and rapid molecular identification
InactiveCN110656196AAccurate amplificationMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationGenetic diversity
The invention discloses a primer group capable of accurately measuring the ITS base sequence of cerasus schneideriana, synthesis and rapid molecular identification. The sequence of the primer group isshown as follows: 1f:5'-CTCCTCGTCCCTTTTCTC-3' (SEQ ID No.1) and 2r:5'-AGTTTCTTTTCCTCCGCT-3' (SEQ ID No.2). The invention relates to a specific pair of primers of 1f-2r, the primers can be used to acquire the ITS sequence of the cerasus schneideriana with good quality through PCR, and the 1f-2r is specific sequence primers for the cerasus schneideriana and used for ITS molecular sequence auxiliaryidentification by combining morphological classification; and in addition, the specific primer pair of 1f to 2r can accurately amplify the ITS sequence of the cerasus schneideriana and can provide acertain basis for population genetic diversity, phylogenesis, rapid molecular identification and the like of the cerasus schneideriana.
Owner:NINGBO CITY COLLEGE OF VOCATIONAL TECH
RT-PCR primer and method for amplifying estrogen receptor alpha genes in female chinemys reevesii
InactiveCN107653302ARapid expansionAccurate amplificationMicrobiological testing/measurementDNA/RNA fragmentationTotal rnaGene
The invention discloses a RT-PCR primer and method for amplifying estrogen receptor alpha genes in female chinemys reevesii. The RT-PCR primer comprises a primer pairs as shown in SEQ ID No:1 and SEQID No:2. The method comprises the following steps: 1) extracting total RNA of a to-be-amplified sample; 2) reversely transcribing the extracted total RNA into cDNA; 3) amplifying the cDNA by using theRT-PCR primer; PT-PCR primer is the PT-PCR primer. According to the invention, a set of RT-PCR primers are designed, the total RNA of the to-be-amplified sample is extracted, and then reverse transcription is conducted to obtain the cDNA. The cDNA is amplified by using the RT-PCR primer. Under the premise of cost reducing, a great number of estrogen receptor alpha genes in female chinemys reevesii fallopian tubes are amplified rapidly and accurately, and large-scale and more intuitive studies of the estrogen receptor alpha genes in the female chinemys reevesii are effectively achieved.
Owner:ANHUI NORMAL UNIV
A novel coronavirus nucleic acid detection kit
ActiveCN114032340BAvoid pollutionReduce exposureMicrobiological testing/measurementAgainst vector-borne diseasesNucleic acid detectionGenome
The invention discloses a novel coronavirus nucleic acid detection kit, which belongs to the technical field of biological detection. The present invention uses the real-time fluorescence quantitative cross primer isothermal amplification technology to detect the novel coronavirus, and designs two sets of primers suitable for this technology. The two sets of primers are respectively for the detection of the ORF1ab gene in the type coronavirus genome and the S gene in the type coronavirus genome. The novel coronavirus nucleic acid detection kit of the invention has strong specificity and high sensitivity, not only can qualitatively and quantitatively detect samples, but also can observe changes in data in real time, which is conducive to the comprehensive collection of sample data and facilitates further research. Therefore, the present invention is not only suitable for on-site rapid detection, but also for detection for the purpose of scientific research.
Owner:HANGZHOU ANYU TECH CO LTD
A dna barcode and its use in identifying muscadine grapes
ActiveCN107779521BVersatileImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMolecular identification
The invention discloses a DNA barcode, the nucleotide sequence of which is shown in SEQ ID NO.1. The DNA barcode disclosed the ITS sequence of Muscadine grape for the first time, which provided an effective molecular marker for the species identification of Muscadine grape. In addition, the DNA barcode also included some conserved sequences, which is universal when applied to the species identification of grapevine. The invention discloses a method for preparing the nucleotide sequence of the DNA barcode and primers, which can be used to obtain the sequence of the ITS region of muscadine grapes. The invention also discloses a molecular identification method of muscadine grapes, detection primers and a kit for identifying muscadine grapes, using the detection primers or kits to carry out PCR amplification to obtain the sequence of the grape ITS2 region, and then combining the amplified sequence with the muscadine The DNA barcode sequences of grapes are compared or applied to construct a phylogenetic tree of grapes to achieve accurate identification of grape varieties, and the identification process is simple, efficient and repeatable.
Owner:NANJING XIAOZHUANG UNIV
A kind of primer set and its application and kit in amplifying siv/shiv genome
ActiveCN105624153BIncreased sensitivityAmplification is stable and accurateMicrobiological testing/measurementDNA preparationSimian human immunodeficiency virusSequence variation
Owner:INST OF LAB ANIMAL SCI CHINESE ACAD OF MEDICAL SCI +1
Detection method of turbot FF0922 microsatellite marker by utilizing specific primers
InactiveCN101705296BEasy to detectEasy accessMicrobiological testing/measurementNegative strandGenomic DNA
The present invention discloses a detection method of a turbot FF0922 microsatellite marker by utilizing specific primers, comprising the following steps: firstly extracting turbot genomic DNA; then utilizing a sequence containing a microsatellite in an enrichment library of the turbot microsatellite; designing specific primers at two ends of the sequence as a positive strand 5'-TGTGAGAAAACAGGCAATC-3', and a negative strand CAGTTTCTGTGATTTGGTGAT-3'; carrying out PCR amplification on the turbot genomic DNA; separating the obtained PCR product through modified polyacrylamide gel electrophoresis; analyzing strips generated on the product to obtain a turbot genetic polymorphism map. The method of the invention is suitable for the detection technologies of turbot population genetic marker, genealogy authentication, genetic linkage map construction and the like; the PCR amplification product presents better polymorphism in turbot population detection; and the method of the invention can rapidly acquire a genetic variation map of the turbot FF0922 microsatellite marker, and conveniently, rapidly, accurately and intuitively detect each individual genotype of the turbot.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
A kind of primer and method for detecting single nucleotide polymorphism of goat dqa1 gene exon 4
ActiveCN104450880BAccurate amplificationAccurate analysisMicrobiological testing/measurementDNA/RNA fragmentationSingle nucleotide mutationAllele
The invention discloses a primer and method for detecting the mononucleotide polymorphism of a DQA1 gene related to goat immunity. The primer comprises an upstream primer and a downstream primer. When detection is carried out, goat genome DNA is taken as a template, PCR (polymerase chain reaction) is carried out by utilizing the upstream primer and the downstream primer to amplify a target fragment, a PCR product is treated by using a denaturing agent, the denatured amplified fragment is subjected to non-denatured polyacrylamide gel electrophoresis detection, glue dyeing is carried out by applying a silver dyeing method, and an allelic gene of a goat DQA1 gene exon 4 can be judged according to a polyacrylamide gel electrophoresis result. According to the nucleotide primer disclosed by the invention and an established detection system, different allelic genotypes of a group can be more accurately and faster amplified, and different mononucleotide mutant sites of the goat DQA1 gene exon 4 can be obtained through accurate analysis.
Owner:广西平果润民发展有限公司
Soybean cleistogamous flower molecular marker and application thereof
InactiveCN111663000AHigh sensitivityImprove featuresMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyGene flow
The invention discloses a soybean cleistogamous flower molecular marker. The molecular marker comprises a DNA molecule SNP2 obtained by using soybean genome DNA as a template and performing amplification by using a SNP2 primer pair, and a DNA molecule SNP3 obtained by using the soybean genome DNA as a template and performing amplification by using a SNP3 primer pair, wherein the SNP2 primer pair and the SNP3 primer pair are as shown in SEQ ID NO.1-SEQ ID NO.4. A method of map-based cloning is used to locate a cleistogamous flower trait decisive base sequence in cleistogamous flower soybean genome, and perform cloning, according to the cloning result, whether the trait of soybean is the cleistogamous flower is determined, and then the soybean is planted, so that the plant maintains a pure breed by avoiding the interference of foreign pollens, and the gene flow is controlled.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Detection method of turbot FF0901 microsatellite marker by utilizing specific primers
InactiveCN101705295BEasy to detectEasy accessMicrobiological testing/measurementNegative strandGenomic DNA
The invention discloses a detection method of a turbot FF0901 microsatellite marker by utilizing specific primers, comprising the following steps: firstly extracting turbot genomic DNA; then utilizing a sequence containing a microsatellite in an enrichment library of the turbot microsatellite; designing specific primers at two ends of the sequence as a positive strand 5'-TTACGCTTCGCATCGCTCAT-3', and a negative strand 5'-TGCCAGGCCACCAGACG-3'; carrying out PCR amplification on the turbot genomic DNA; separating the obtained PCR product through modified polyacrylamide gel electrophoresis; analyzing strips generated on the product to obtain a turbot genetic polymorphism map. The method of the invention is suitable for the detection technologies of turbot population genetic marker, genealogy authentication, genetic linkage map construction and the like; the PCR amplification product presents better polymorphism in turbot population detection; and the method of the invention can rapidly acquire a genetic variation map of the turbot FF0901 microsatellite marker, and conveniently, rapidly, accurately and intuitively detect each individual genotype of the turbot.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Viral hepatitis treatment drug interferon production technology
InactiveCN104530213AImprove stabilityHigh activityPeptide/protein ingredientsDigestive systemFreeze-dryingInterferon production
The invention discloses a viral hepatitis treatment drug interferon production technology which comprises the following steps: (1) acquisition of a target gene; (2) amplifying by PCR (polymerase chain reaction), to be more specific, in a microcentrifuge tube, using the 1 g / mL target gene obtained by the step (1) as template DNA, adding a right amount of a buffer solution, 4 kinds of dNTP (diethyl-nitrophenyl thiophosphate) mixtures, 0.5 2.5U / 50 muL DNA polymerase system, a pair of 0.1-0.5 mumol / L synthesized DNA primer to make the system pH 6.8-7.8; (3) gene recombination; (4) transformation; (5) separation and purification; (6) gene expression; (7) modification; and (8) freeze-drying, mass viral hepatitis treatment drug interferon can be produced in a short period of time, after modifying with polyethylene glycol, the half-life of the interferon is prolonged, hepatitis virus targeting property is improved, the antigenicity is weak, no human body immune response is caused, drug efficacy is good, production is fixed, stability is good, and the viral hepatitis treatment drug interferon production technology is suitable for popularization and application.
Owner:GUANGXI UNIV
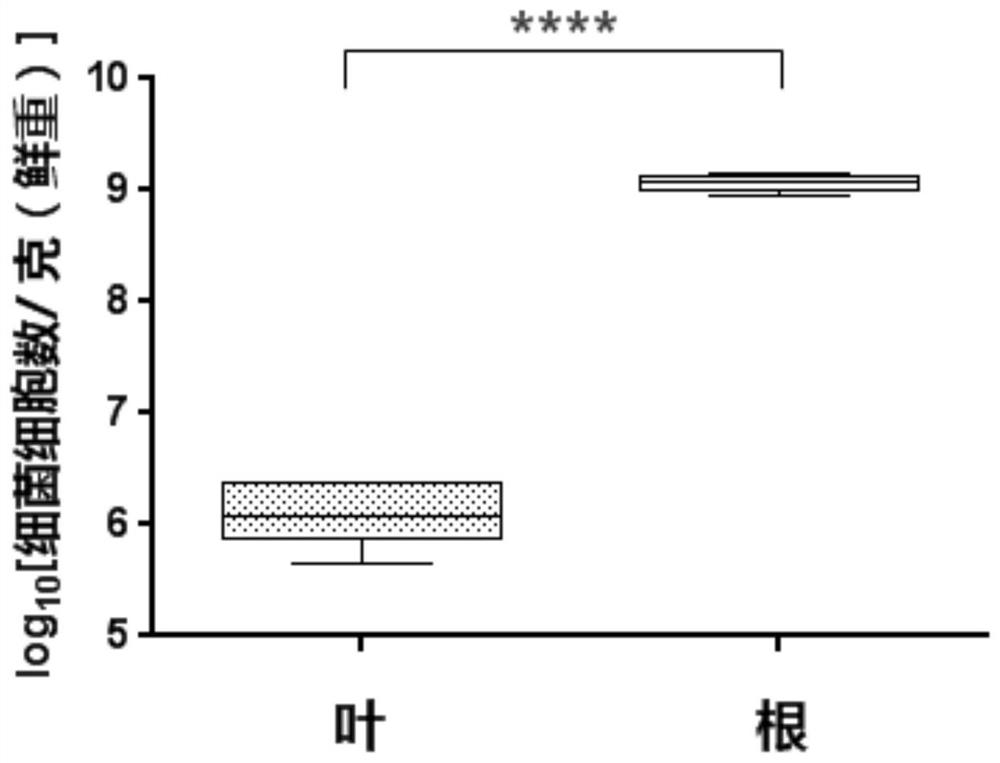
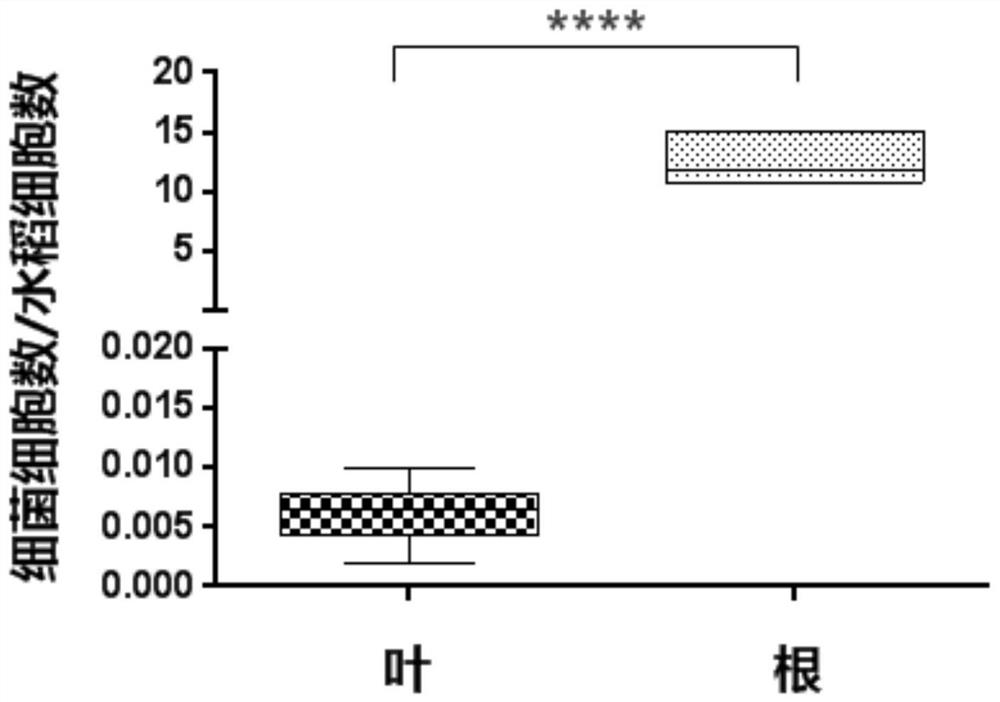
Method for quantitatively detecting endophytic bacteria of plant tissue by non-culture method
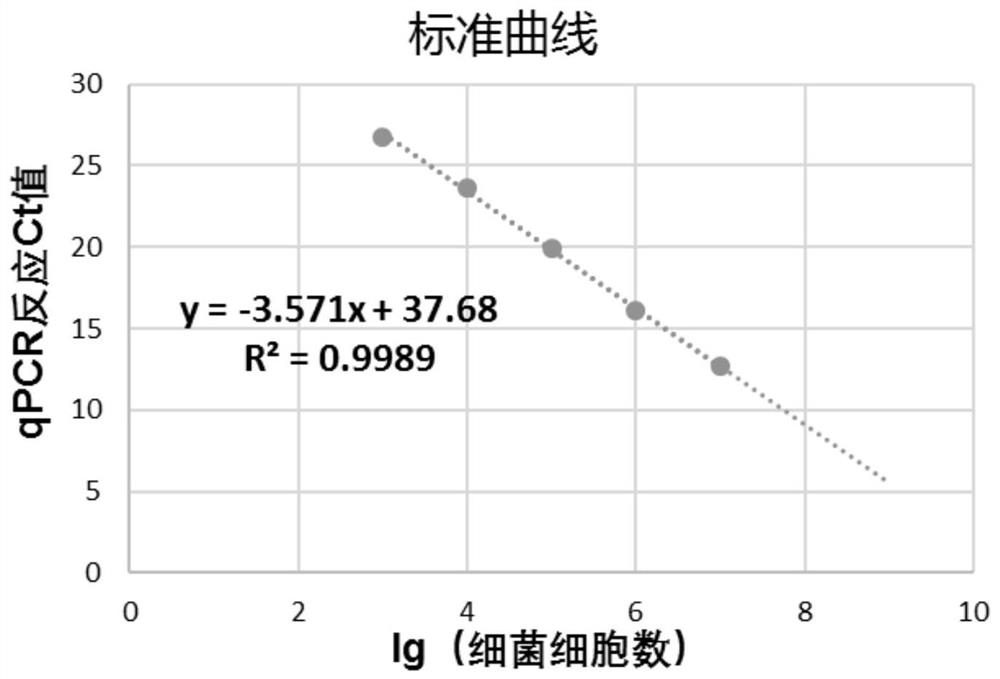
The invention discloses a method for quantitatively detecting endophytic bacteria of plant tissues through a non-culture method. The method comprises the following steps: adding bacterial DNA with different copy numbers into genome DNA extracted from sterile calluses, and carrying out fluorescent quantitative PCR (Polymerase Chain Reaction) by using a bacterial 16S rRNA gene specific primer pair to obtain a standard curve of the plant tissue bacterial content and a fluorescent quantitative PCR reaction Ct value; carrying out surface sterilization on plant tissues to be detected, and extracting total DNA; and taking the extracted DNA as a template, carrying out fluorescent quantitative PCR (Polymerase Chain Reaction) by adopting the bacterial 16S rRNA gene specific primer pair, and substituting the obtained Ct value into the standard curve to obtain the content of the endophytic bacteria in the plant tissue to be detected. The method is easy and convenient to operate, time-saving, efficient, capable of covering all endophytic bacteria of plant tissues and suitable for various plants, and the bacterial content of the plant tissues to be detected can be directly obtained by substituting the qPCR reaction Ct value of a sample to be detected into the standard curve in related research.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
A primer set, kit and detection method for rapid detection of human dys385 gene
ActiveCN110791557BShort detection timeImprove detection accuracyMicrobiological testing/measurementDNA/RNA fragmentationGeneBioinformatics
The invention discloses a primer set for rapidly detecting human DYS385 gene. Wherein, the primer set is composed of the following primers: primer F1 as shown in SEQ ID No.1; primer B1 as shown in SEQ ID No.2; primer F2-1 as shown in SEQ ID No.3; primer B2-1 As shown in SEQ ID No.4; Primer F2-2 as shown in SEQ ID No.5; Primer B2-2 as shown in SEQ ID No.6; Primer LOOP-F as shown in SEQ ID No.7; Primer LOOP -B is shown in SEQ ID No.8. The detection time required by the present invention is short, and the corresponding primers and kit can quickly detect the human DYS385 gene within 30 minutes to determine the sex of the human sample.
Owner:THE FIRST RES INST OF MIN OF PUBLIC SECURITY +1
A primer and kit for detecting bacterial macrolide drug resistance genes
ActiveCN104561341BRapid expansionAccurate amplificationMicrobiological testing/measurementDNA/RNA fragmentationBacteroidesMicroorganism
The invention discloses primers and a kit for detecting the macrolide drug-resistance genes of bacteria, and belongs to the technical field of microbiological detection. The sequences of the primers for detecting the macrolide drug-resistance genes of bacteria provided by the invention are shown in SEQ ID NO: 1-6, and the three primer pairs are capable of specifically amplifying the three macrolide drug-resistance genes of ermA, ermB and ermC of multiple bacteria. The invention further discloses a kit for detecting the macrolide drug-resistance genes of bacteria, wherein the kit contains an EvaGreen fluorescent dye, and is capable of rapidly and accurately detecting the macrolide antibiotic-related drug-resistance genes of bacteria. The primers and the kit disclosed by the invention have the advantages of simplicity and convenience in operation, high specificity, high sensibility, low cost, high flux and the like, and can be used for rapidly detecting macrolide antibiotic-related drug-resistance genes of clinic pathological bacteria and providing a reference for clinic treatment.
Owner:CHONGQING DIAN SRAB CENT FOR CLINICAL LAB CO LTD
Scophthalmus maximus T170G single nucleotide polymorphic marking detection method
InactiveCN102586454BAccurate amplificationGood polymorphismMicrobiological testing/measurementScophthalmusGenotype
The invention relates to a scophthalmus maximus T170G single nucleotide polymorphic marking detection method. The method comprises the following steps: extracting scophthalmus maximus genome DNA and diluting for later use; analyzing the sequences of EST, screening the sequence containing a candidate SNP locus, designing a specific primer at the two ends and designing a non-marking probe with the closed 3' end in front of and behind the locus (comprising the locus); performing asymmetric PCR amplification on the scophthalmus maximus group genome DNA by using the primer; hybridizing the amplification product with the non-marking probe with the closed 3' end; and placing the hybrid product on a LightScanner, and detecting and analyzing the melting curve to obtain the genetic polymorphism map of the scophthalmus maximus. The method is applicable to detection technologies of scophthalmus maximus genetic marking, genealogy authentication, genetic linkage map construction and the like. According to the method which is convenient, quick and accurate, the scophthalmus maximus T170GSNP marked genetic variation map can be obtained quickly; and the genotype of each individual of the scophthalmus maximus can be detected intuitively.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Poppy Species Specific Genetic Marker Detection System
ActiveCN105861711BAccurate amplificationRapid amplificationMicrobiological testing/measurementDNA/RNA fragmentationCorrosionPoppy
The invention discloses a poppy species specificity genetic marker detecting system. A primer pair set for identifying poppy is provided and is composed of a primer pair P12 (sequence 1 and 2), a primer pair P13 (sequence 3 and 4), a primer pair P5 (sequence 5 and 6) and a primer pair P14 (sequence 7 and 8). Four STR loci of poppy are amplified at the same time, and the amplification is more accurate and rapid compared with amplification of a single locus; the system has higher species specificity when identifying poppy and affinis plants, and an effective method is provided for further studying poppy STR loci and inspecting and identifying the poppy species in a case; in an optimized poppy STR composite amplification system, amplification of all loci can be balanced, sensitivity is high, stability is high, and frequently-seen corrosion check materials in a drug-related case can be easily inspected; accuracy is high, parting of four loci of each of poppy samples from different producing areas can be achieved through detection and data statistic analysis.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Kit for detecting susceptibility gene of liver cancer of HBeAg negative HBV chronic infection liver cirrhosis patient and method
ActiveCN106834455AStrong specificityAccurate amplificationMicrobiological testing/measurementTLR9Hbeag negative
The invention provides a kit for detecting wherein an HBeAg negative HBV chronic infection liver cirrhosis patient is susceptible population to liver cancer or not and a method and relates to the field of biodetection. The kit comprises an amplification reaction liquid for amplifying a specific primer of DNA containing an SNP site in a sample, a purifying reaction liquid for purifying SAP enzyme of the DNA and an extending reaction liquid of a single basic group extending primer for obtaining a basic group detection result at rs187084 site of an TLR9 gene; when the detection result at rs187084 site of the TLR9 gene appears as a CT or / and TT genotype, the HBeAg negative HBV chronic infection liver cirrhosis patient is susceptible population to liver cancer. The specific primer and the extending primer provided by the invention can accurately amplify and extend a target gent, are high in sensitivity and good in specificity and can provide a ground for judging whether the HBeAg negative HBV chronic infection liver cirrhosis patient is developed to hepatocellular carcinoma or not, and the method is simple to operate, and a prognostic target is favorably monitored.
Owner:PEKING UNIV FIRST HOSPITAL +1
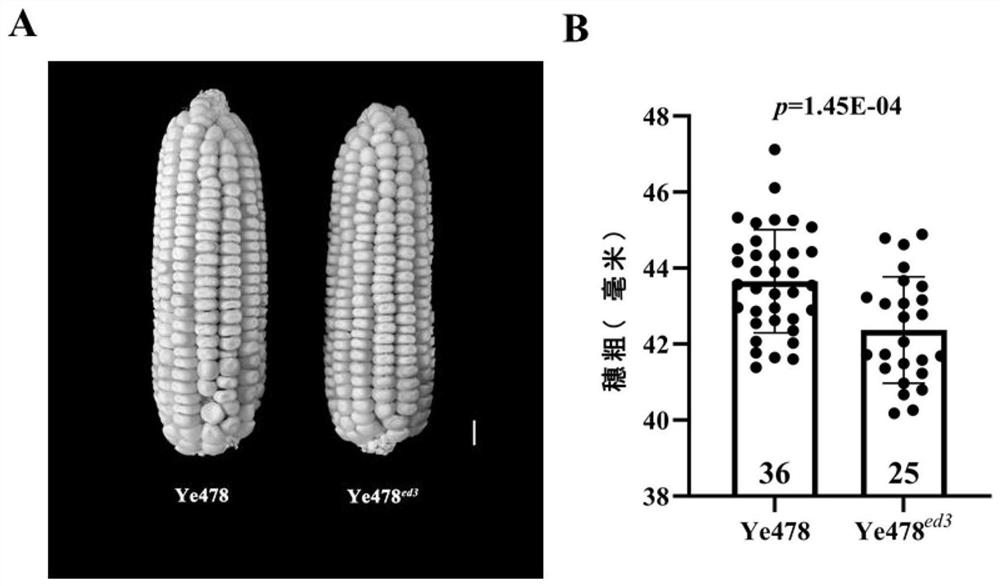
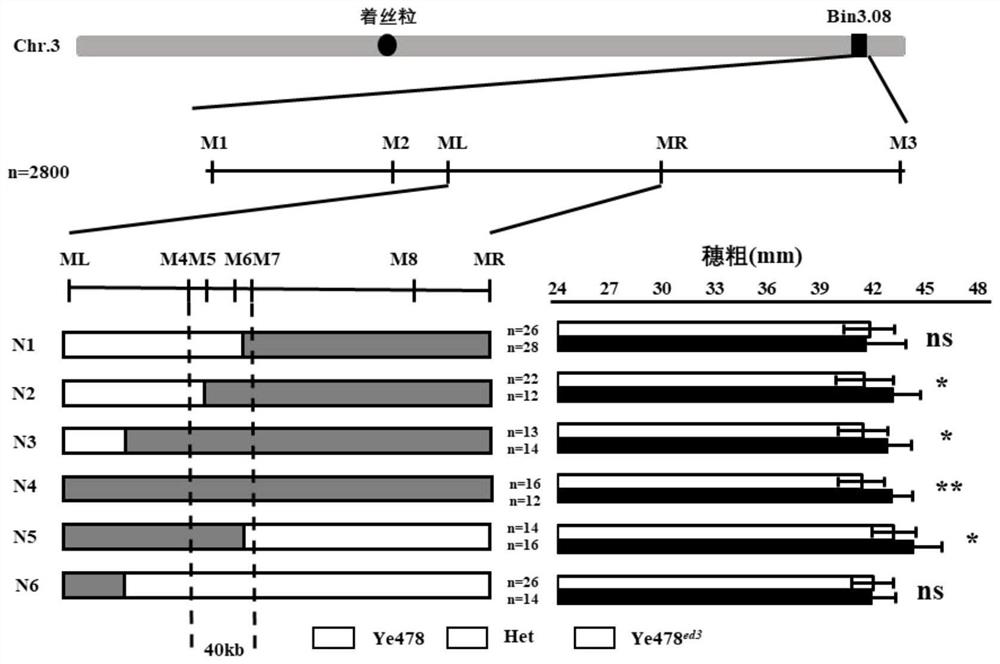
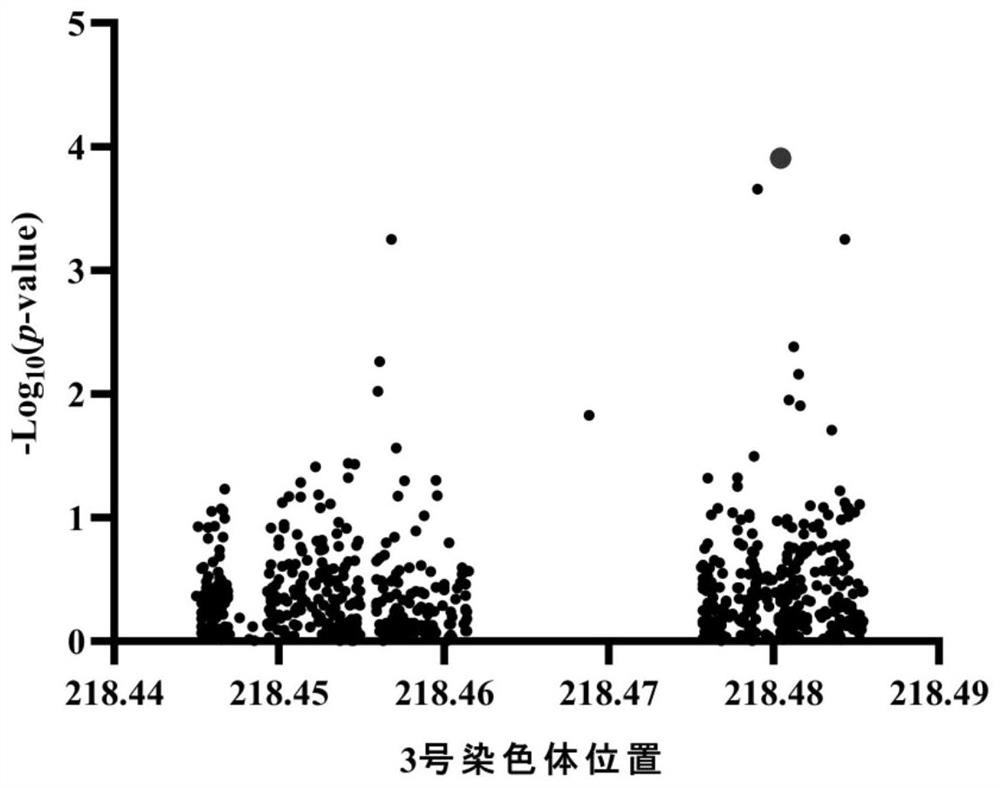
Pair of molecular marker primers closely linked with corncob coarse QTL ZmED3 and application thereof
ActiveCN112646913AAccurate amplificationStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationNucleotideNucleotide sequencing
The invention belongs to the technical field of biology, and particularly relates to a pair of molecular marker primers closely linked with corncob coarse QTL ZmED3 and application thereof. The invention provides the pair of molecular marker primers closely linked with the corncob coarse QTL ZmED3, an upstream primer of the molecular marker primers is shown as SEQ ID NO:1, and a downstream primer of the molecular marker primers is shown as SEQ ID NO:2. The molecular marker primers have a close linkage relationship with a gene for controlling the coarse character of corncobs, can be used for accurately amplifying to obtain a nucleotide sequence shown as SEQ ID NO:3 or SEQ ID NO:4, and have the characteristic of high amplification specificity. The molecular marker primers can effectively achieve the technical effect of identifying the corncob thick character, and can be effectively applied to genetic improvement of the corncob thick character.
Owner:HUAZHONG AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com