A kind of primer set and its application and kit in amplifying siv/shiv genome
A primer amplification and genome technology, applied in the field of molecular biology, can solve problems such as sequence analysis troubles, reduced accuracy, sequence selection and limited amplified sequences, and achieve good sensitivity results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Using the SIVmac239 plasmid as a template, the primers listed in Table 1 were amplified.
[0067] Table 1 Primer Sequence
[0068]
upstream
downstream
Amplified fragment length
primer pair 1
SEQ ID NO:1
SEQ ID NO:2
5728bp
primer pair 2
SEQ ID NO:9
SEQ ID NO:10
5398bp
primer pair 3
SEQ ID NO:9
SEQ ID NO: 11
5381bp
primer pair 4
SEQ ID NO:3
SEQ ID NO:4
5103bp
primer pair 5
SEQ ID NO:12
SEQ ID NO: 13
5022bp
Primer pair 6
SEQ ID NO: 14
SEQ ID NO:6
4811bp
primer pair 7
SEQ ID NO:5
SEQ ID NO:6
4842bp
Primer pair 8
SEQ ID NO:7
SEQ ID NO:8
4766bp
[0069] The amplification system is:
[0070]
[0071] The amplification procedure is:
[0072]
[0073]
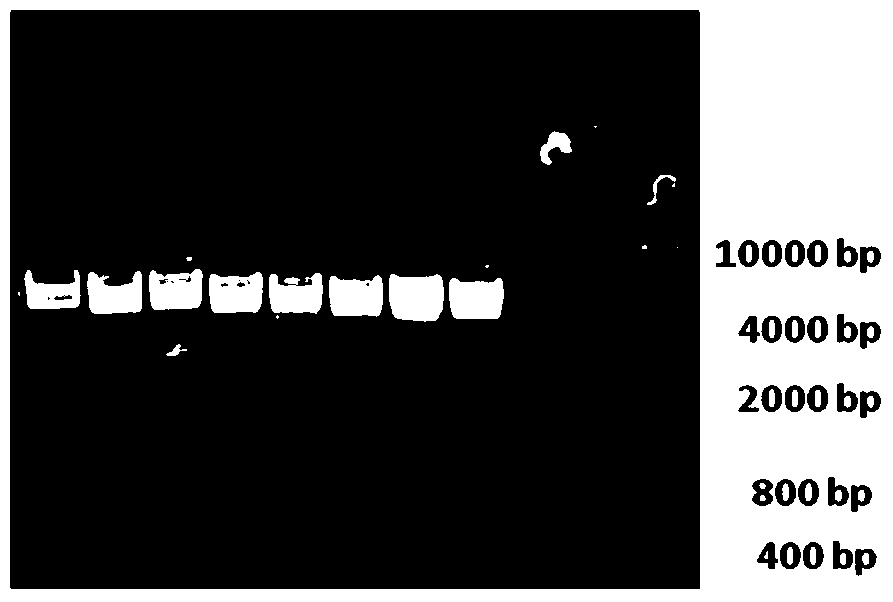
[0074] The amplified product was subjected to 1.2% TAE agarose gel electrophoresis, and the results were as follows: figure 1 , wherein, lanes...
Embodiment 2
[0076] The primers in Table 2 were used to amplify by means of nested PCR. The first round of amplification used the culture supernatant of SIVmac239 as a template, and the second round of amplification used the amplification product of the first round as a template. Experimental setup Two experimental groups were used to amplify the 3' half-length and 5' half-length of the genome respectively. Another 10 comparison groups were set up.
[0077] Table 2 Primer Sequence
[0078]
[0079]
[0080] The amplification system is:
[0081]
[0082] The amplification procedure is:
[0083]
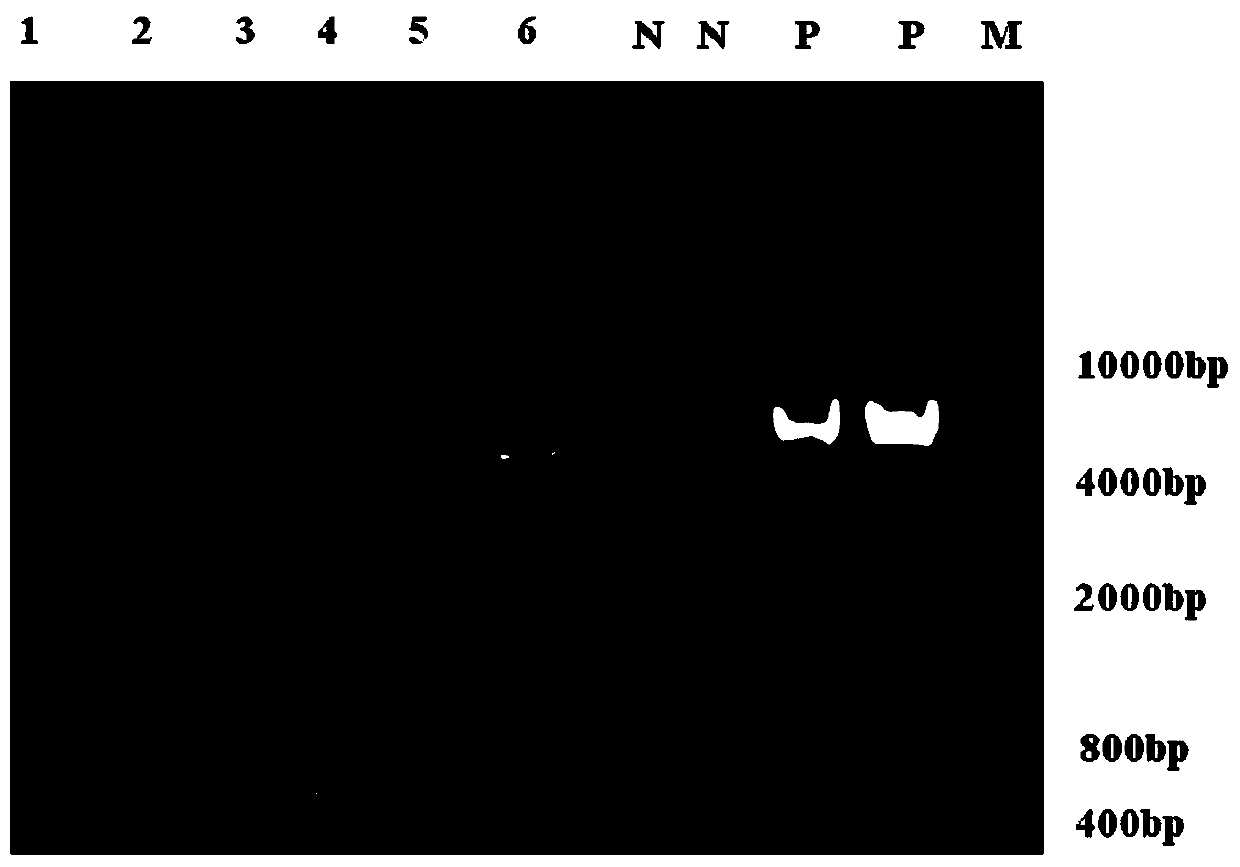
[0084] After two rounds of PCR amplification in each group of experiments, the products were subjected to 1.2% TAE agarose gel electrophoresis, and the results were as follows: Figure 2-a and Figure 2-b . It can be seen from the figure that the primer pair provided by experimental group 1 is most suitable for amplifying the 5' end of the SIV genome, while the primer pair provided...
Embodiment 3
[0087] The primers in Table 2 in Example 2 were used to amplify by means of nested PCR. The first round of amplification used the SIVmac239 plasmid as a template, and the second round of amplification used the amplification product of the first round as a template. Experimental setup Two experimental groups were used to amplify the 3' half-length and 5' half-length of the genome respectively. Another 10 comparison groups were set up.
[0088] The amplification system is:
[0089]
[0090] The amplification procedure is:
[0091]
[0092] After two rounds of PCR amplification in each group of experiments, the products were subjected to 1.2% TAE agarose gel electrophoresis, and the results were as follows: Figure 3-a and Figure 3-b . It can be seen from the figure that the primer pair provided by experimental group 1 is most suitable for amplifying the 5' end of the SIV genome, while the primer pair provided by experimental group 2 is most suitable for amplifying the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com