A primer set, kit and detection method for rapid detection of human dys385 gene
A detection kit and primer set technology, which is applied in the field of forensic detection, can solve problems such as the limited number of detection personnel, the impossibility of on-site rapid detection of detection materials, and the difficulty of detection, so as to save material costs and time costs, and the results are intuitive and visualized , The effect of low detection environment requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Primer Design
[0049] DNA template (SEQ ID No. 9):
[0050] AGCATGGGTGACAGAGCTAGACACCATGCCAAACAACAACAAAGAAAAGAAATGAAATTCAGAAAGGAAGGAAGGAAGGAGAAAGAAAGTAAAAAAGAAAAAAGAGAAAAAGAGAAAAAGAAAGAAAGAGAAGAAAGAGAAAGAGGAAAGAGAAAGAAAGGAAGGAAGGAAGGAAGGAAGGGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAGAAAAAGAAAGGAGGACTATGTAATTGGAATAGATAGATTATTTTTTAAAATATTTTTATTACCTTTACAGTTTTTTTAAATGCCGCCATTTCAGAAAGAAATCTGGTCACAGCCCTTACCAGCTTTACCTAGCATCCCA
[0051] Primer design: Primers were designed by LAMP online design software https: / / primerexplorer.jp.
[0052] Primer synthesis company: Zhongmeitaihe Biotechnology (Beijing) Co., Ltd.
[0053] The primers screened by the design of the present invention are as follows:
[0054] Table 1
[0055] primer name Primer number Primer sequence Primer F1 SEQ ID No.1 AGCATGGGTGACAGAGCTA Primer B1 SEQ ID No.2 TGGGATGCTAGGTAAAGCTG Primer F2-1 SEQ ID No.3 ACACCATGCCAAACAACAAC Primer B2-1 ...
Embodiment 2
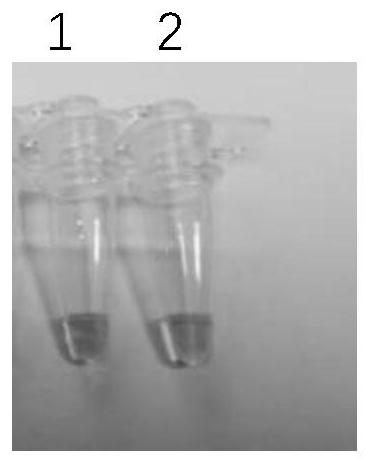
[0056] Embodiment 2 utilizes the test kit of the present invention to detect the accuracy verification
[0057] 1. Sample to be tested: 9948DNA (purchased from promega company)
[0058] 2. Use the kit of the present invention to detect the above-mentioned sample:
[0059] (1) Sample amplification:
[0060] Preparation reaction system: Bst DNA polymerase (8U / ul) 1ul, 10*Bst DNA polymerase buffer 2.5ul, primer F1 (SEQ ID No.1) 10mmol / L 1ul, primer B1 (SEQ ID No.2) 10mmol / L 1ul, primer F2-1 (SEQ ID No.3) 10mmol / L 1ul, primer B2-1 (SEQ ID No.4) 10mmol / L 1ul, primer F2-2 (SEQ ID No.5) 10mmol / L1ul, primer B2 -2 (SEQ ID No.6) 10mmol / L 1ul, primer LOOP-F (SEQ ID No.7) 10mmol / L1ul, primer LOOP-B (SEQ ID No.8) 10mmol / L 1ul, MgSO 4 (100mmol / L) 0.9ul, calcein (625umol / L) 1ul, manganese chloride (7.5mmol / L) 1ul, sample DNA (10ng / ul) 1ul, double distilled water to make up 25ul.
[0061] At the same time, a control group was set up, except that the sample DNA was replaced with double dis...
Embodiment 3
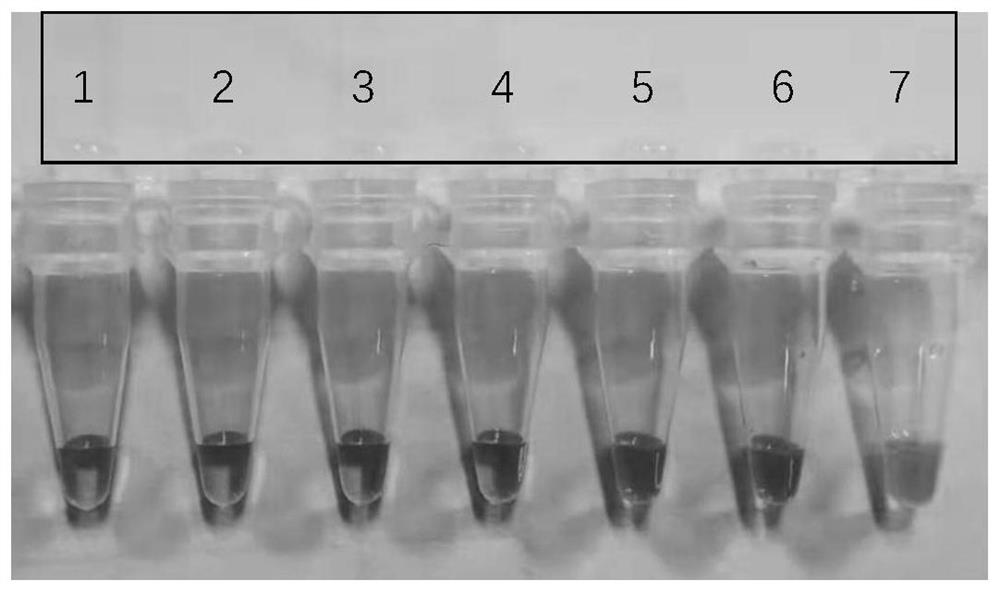
[0064] Example 3 Sample size test of the kit of the present invention
[0065] (1) Sample amplification:
[0066] Sample DNA gradient dilution: use double-distilled water to dilute the concentration of 9948 DNA. The diluted sample concentrations are: 10ng / ul, 5ng / ul, 2ng / ul, 1ng / ul, 0.5ng / ul, 0.2ng / ul, 0.1 ng / ul.
[0067] Preparation reaction system: Bst DNA polymerase (8U / ul) 1ul, 10*Bst DNA polymerase buffer 2.5ul, primer F1 (SEQ ID No.1) 10mmol / L 1ul, primer B1 (SEQ ID No.2) 10mmol / L 1ul, primer F2-1 (SEQ ID No.3) 10mmol / L 1ul, primer B2-1 (SEQ ID No.4) 10mmol / L 1ul, primer F2-2 (SEQ ID No.5) 10mmol / L1ul, primer B2-2 (SEQ ID No.6) 10mmol / L 1ul, primer LOOP-F (SEQ ID No.7) 10mmol / L1ul, primer LOOP-B (SEQ ID No.8) 10mmol / L 1ul, MgSO 4 (100mmol / L) 0.9ul, calcein (625umol / L) 1ul, manganese chloride (7.5mmol / L) 1ul, diluted sample DNA 1ul, double distilled water to make up 25ul. Wherein PCR tubes 1 to 7 correspond to DNA templates of 10ng / ul, 5ng / ul, 2ng / ul, 1ng / ul, 0.5ng / u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com