Kit for detecting susceptibility gene of liver cancer of HBeAg negative HBV chronic infection liver cirrhosis patient and method
A chronic infection and kit technology, applied in the medical and health field, can solve the problem of no clinical prediction, no relationship between clinical outcomes, etc., and achieve monitoring that is conducive to prognostic goals, high sensitivity, and good specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Kit
[0048] 1. Amplification reaction solution
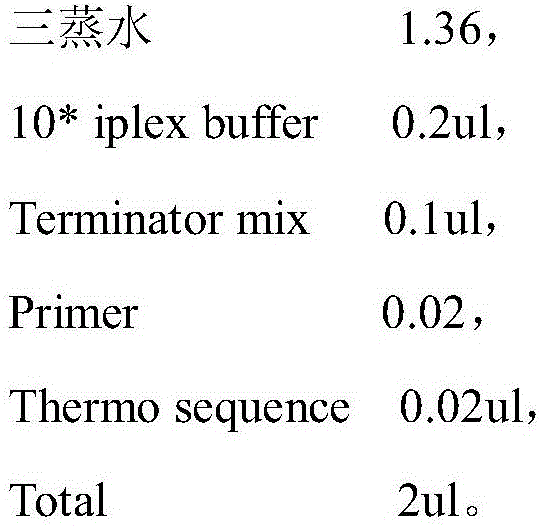
[0049] Used to amplify DNA containing SNP sites in samples, including: 10*buffer 0.625ul, Mg 2+ 0.325ul, dNTP0.25ul, Forward Primer lul, Reverse Primer 1ul, DNA 1ul (10ng-20ng), Hotstar taq enzyme 0.2ul.
[0050] Wherein, the Forward Primer (ie forward primer) is:
[0051] 5'-ACGTTGGATGTATTCCCCCTGCTGGAATGTC-3'.
[0052] Wherein, the Reverse Primer (ie reverse primer) is:
[0053] 5'-ACGTTGGATGTTACTATGTGCTGGGCACTG-3'.
[0054] Further, the reaction conditions for using the amplification reaction solution are: 95°C for 15 minutes; 45 cycles of 94°C for 20s, 56°C for 30s, 72°C for 60s, 72°C for 3min, and storage at 10°C.
[0055] 2. Purify the reaction solution
[0056] Used to purify the amplified DNA fragment, its main component is SAP enzyme, specifically: triple distilled water 1.53ul, Buffer0.17ul, SAP 0.3ul.
[0057] Further, the reaction conditions for using the purified reaction solution are: 37°C f...
Embodiment 2
[0062] Example 2 Method for Detecting Whether HBeAg-negative Patients Are Susceptible to Liver Cancer
[0063] 1. Specimen collection and DNA extraction
[0064] Take 200 μl of the subject's blood clot (the blood clot is dissolved in TES solution), and use the QIAGEN QIAampDNA BloodMini Kit to extract the whole genome DNA according to the operating instructions.
[0065] The specific operation steps are as follows:
[0066] 1) First add 200 μl of AL buffer into a 1.5ml centrifuge tube. Then the patient's specimen was gently blown and mixed, and 200 μl of the specimen was drawn into a centrifuge tube containing lysis buffer (AL buffer).
[0067] 2) Add 20 μl of QIAGEN protease or proteinase K to the centrifuge tube in step 1), shake and mix for 15 seconds, centrifuge at 8000 rpm / min for 10 seconds, and then put it into a 56°C electric thermostat water bath for 10 minutes.
[0068] 3) Centrifuge at low speed so that the liquid in the cap of the centrifuge tube enters the cent...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com