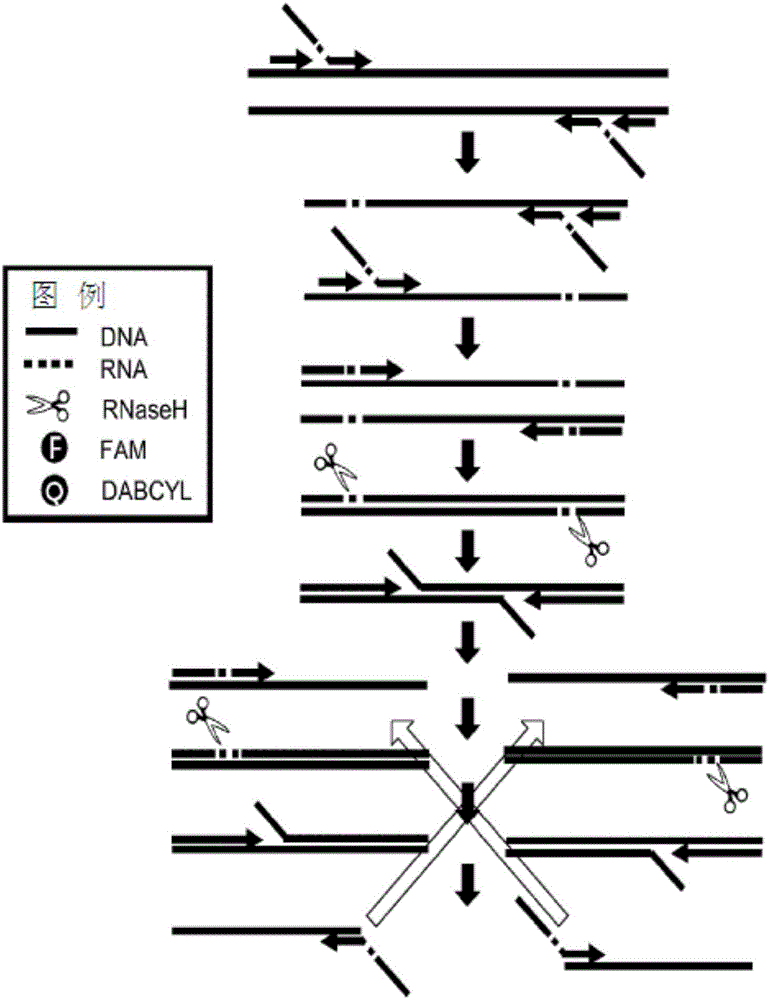
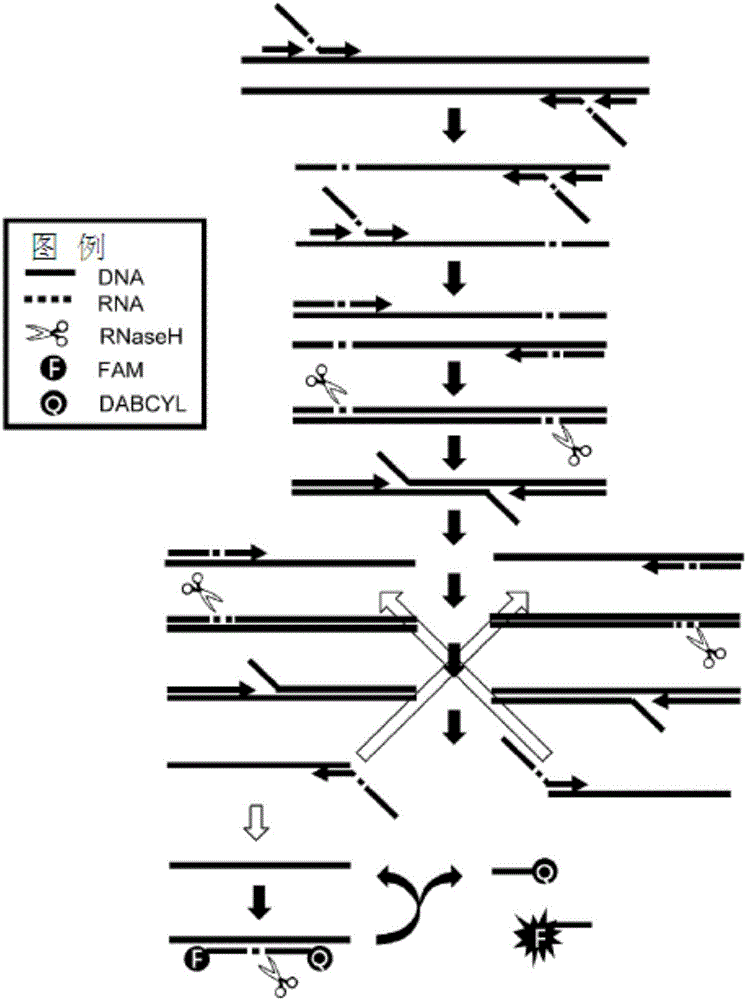
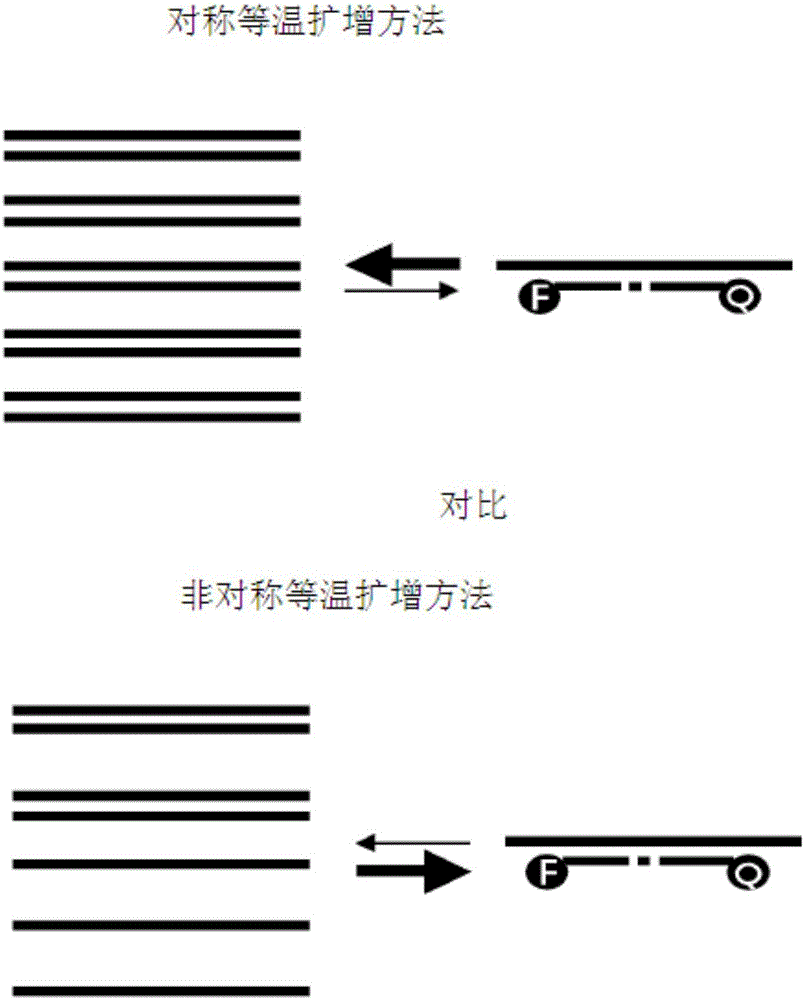
Method for detecting nucleic acid using asymmetric isothermal amplification of nucleic acid and signal probe
A detection method and isothermal amplification technology, applied in the field of needles, can solve problems such as false positive results and limitations, and achieve the effects of rapid and accurate amplification, accurate diagnosis, and no risk of contamination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1: Isothermal amplification of tuberculosis DNA
[0067] As the target DNA, commercially available Mycobacterium tuberculosis DNA group DNA was quantitatively diluted and used as a template for the isothermal amplification reaction. (Vircell, Spain)
[0068] The external primers (SEQ ID NO: 1 and SEQ ID NO: 2) were designed to include a sequence complementary to the nucleotide sequence of Mycobacterium tuberculosis IS6110, and SEQ ID NO: 1 (forward, sense) and SEQ ID NO: 2 (reverse, anti-sense) are as follows.
[0069] Serial number 1: 5'-CGATCGAGCAAGCCA-3'
[0070] Serial number 2: 5'-CGAGCCGCTCGCTGA-3'
[0071] In the DNA-RNA-DNA hybrid primers (SEQ ID NO: 3 and SEQ ID NO: 4), the oligomeric DNA-RNA part at the 5' end has a sequence non-complementary to the base sequence of Mycobacterium tuberculosis IS6110, and the oligomeric DNA part at the 3' end It has a sequence complementary to the nucleotide sequence of Mycobacterium tuberculosis IS6110, and the sequ...
Embodiment 2
[0079] Example 2: Isothermal amplification of Chlamydia DNA
[0080] As target DNA, Chlamydi Atrachomatis DNA group DNA was used. In order to extract the DNA group DNA of Chlamydia, in the Chlamydia strain (ATCC Cat.No.VR-887), use the G-spinTM DNA group DNA extraction kit (iNtRON Biotechnology, Cat.No.17121), after the DNA group DNA is extracted, Perform the amplification reaction. When extracting the DNA group DNA, centrifuge 500 μl of the bacterial suspension at 13,000 rpm for 1 minute and remove the supernatant, hybridize a small amount of 500 μl of phosphate buffer saline (PBS) (pH 7.2) for centrifugation and remove the supernatant, add 300 μl of G-buffer solution of RNase A and Proteinase K were used to suspend the cell pellet and stand at 65° C. for 15 minutes. Next, 250 µl of binding buffer (Binding buffer) was added to uniformly hybridize, and then the spin column was bound to the DNA. After adding 500 ml of washing buffer (washing buffer) A, it was centrifuged at ...
Embodiment 3
[0092] Example 3: Isothermal Amplification of Neisseria gonorrhoeae DNA
[0093] As the target DNA, NeisseriA gonorrhoease DNA group DNA was used. In order to extract DNA group DNA of Neisseria gonorrhoeae strain (ATCC Cat. No. 49226), use G-spinTM DNA group DNA extraction kit (iNtRON Biotechnology, Cat. No. 17121), after extracting DNA group DNA, perform amplification reaction. When extracting the DNA group DNA, centrifuge 500 μl of the bacterial suspension at 13,000 rpm for 1 minute and remove the supernatant, hybridize a small amount of 500 μl of phosphate buffer saline (PBS) (pH 7.2) for centrifugation and remove the supernatant, add 300 μl of G-buffer solution of RNase A and Proteinase K were used to suspend the cell pellet and stand at 65° C. for 15 minutes. Next, 250 µl of binding buffer (Binding buffer) was added to uniformly hybridize, and then the spin column was bound to the DNA. After adding 500 ml of washing buffer (washing buffer) A, it was centrifuged at 1300...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com