Soybean cleistogamous flower molecular marker and application thereof
A molecular marker, soybean technology, applied in the direction of DNA/RNA fragment, microbial determination/inspection, recombinant DNA technology, etc., can solve the problem that soybean application has not made substantial progress, achieve excellent sensitivity and specificity, and prevent false positives Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] A method for obtaining molecular markers of soybean atresia, including the following steps:
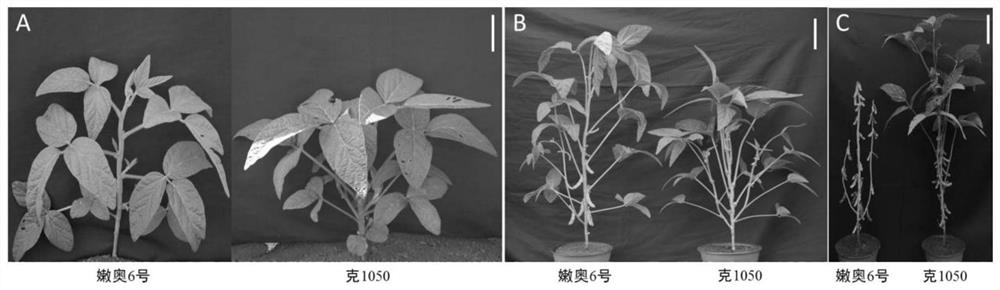
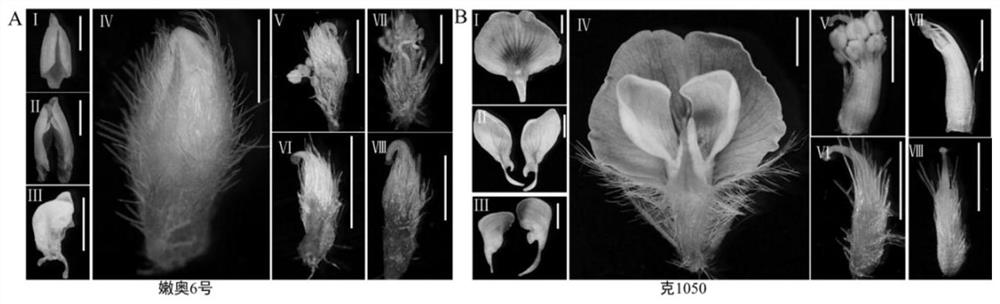
[0038] (1) To construct F with normal flower open soybean variety Ke1050 as the female parent and closed flower variety N639 as the male parent 2 Separated populations; field phenotypic identification results show that Ke1050 is a flower flag petal, which spreads out after pollination, and N639 is a flower flag petal, and the wing petals are closed for pollination. The qualitative statistics (open flower, closed flower) of the flower opening habits of the F2 population are carried out. Genetic analysisχ 2 =1.202 0.05 = 3.84, indicating that the atretic flower phenotype is controlled by a pair of recessive nuclear genes;
[0039] (2) Extract the parent and its F 2 Population leaf genomic DNA; use CTAB method to extract parents and F 2 The genomic DNA of the leaves of the segregated population of generations. Take an equal amount of mixing leaves in a 2mL centrifuge tube, put them int...
Embodiment 2
[0096] The process of identifying soybean atresia flower traits by molecular markers:
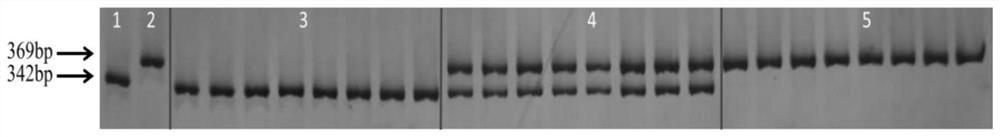
[0097] Extract the genomic DNA of the soybean to be tested, and use the genomic DNA as a template to carry out PCR amplification of SNP2 and SNP3 with primers (PCR reaction program: 95℃5min; 95℃20S, 58℃30S, 72℃30S, 34 cycles ;72℃5min.), take 1μL PCR product, Cutsmart Buffer 1μL, restriction enzyme (20,000U / mL) 0.2μL, add 7.8μL water, the total volume is 10μL, centrifuge and mix, SNP2 digestion at 65℃ for 2h, SNP3 was digested at 37°C for 2h. Then, 6% non-denaturing polyacrylamide gel electrophoresis and silver nitrate staining were performed sequentially. Then make the following judgments:
[0098] Using the genomic DNA of the soybean N639 and grams 1050 to be tested as a template, the specific primer SNP2 is used to perform PCR amplification. If a 342bp fragment is obtained, the flower opening habit of the soybean to be tested is or the candidate is a closed flower, If a 369bp fragment is ob...
Embodiment 3
[0105] Example 3 Primer Sensitivity Detection
[0106] Method: Use primer pair SNP2 and primer pair SNP3 to amplify parental DNA of different concentrations. The DNA concentrations of N639 and g 1050 are set to 100ng / μL, 80ng / μL, 60ng / μL, 40ng / μL, 20ng / μL, 10ng / μL, 5ng / μL, 2ng / μL, 1ng / μL, 0.5ng / μL, 0.2ng / μL, 0.1ng / μL.
[0107] PCR amplification system: DNA at the above concentration, primers (F+R) 1.5μL, Buffer 2μL, dNTP 1.5μL, EasyTaq enzyme 0.2μL, sterile water dilute to 20μL.
[0108] Reaction procedure: 95°C for 5min, 95°C for 20s, 58°C for 30s, 72°C for 30s, 72°C for 5min, 34 cycles.
[0109] The result is Figure 5 with Image 6 Shown.
[0110] It can be seen that the minimum amplification concentration of primer pairs SNP2 and SNP3 is 20ng / μL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com