Camphor tree whole genome ssr molecular marker and its preparation method and application
A molecular marker and genome-wide technology, applied in biochemical equipment and methods, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problem of restricting the research and development of camphor tree molecular biology, difficult to meet the needs of camphor tree EST-SSR marker development, Problems such as lack of camphor tree EST data
Active Publication Date: 2022-01-18
JIANGXI ACAD OF FORESTRY
View PDF2 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
However, compared with other species such as rice, corn, grapes, etc., the EST data of camphor tree included in the database of the National Center for Biotechnology Information (NCBI) is quite lacking, which is difficult to meet the needs of camphor tree EST-SSR marker development, and cannot be used for the development of camphor tree EST-SSR markers. DNA molecular marker development research provides sufficient information
This also greatly limits the development of camphor tree molecular biology research
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 1
[0077] One, the preparation method of the primer based on the SSR molecular marker of camphor tree genome development, comprises the following steps:
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
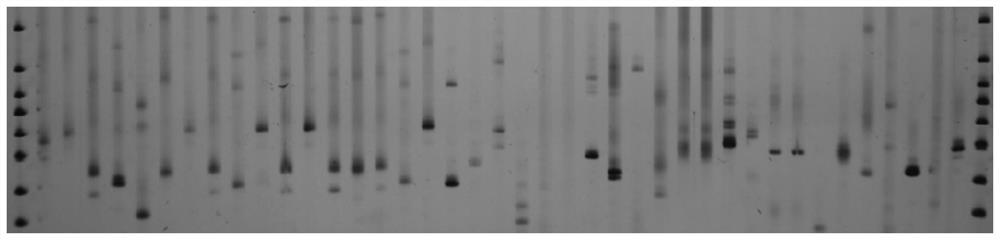
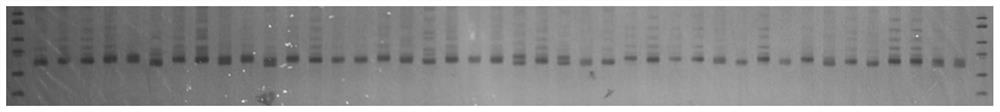
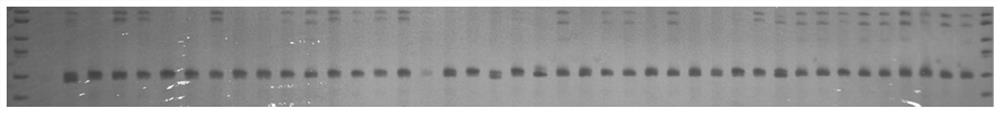
The invention discloses a whole genome SSR molecular marker of camphor tree and its preparation method and application. The camphor tree microsatellite molecular markers developed by the present invention can be applied to the population genetic diversity detection and kinship analysis of ancient camphor trees, providing a new batch of microsatellite molecular markers for Lauraceae plants, and also for the population genetic differentiation of Lauraceae plants New tools are provided for the study of genetic diversity and structure, the level of genetic diversity of populations, the gene flow and mating system between populations, species evolution, and molecular assisted breeding.
Description
Technical field: [0001] The invention belongs to the technical field of forestry molecular biology, and specifically relates to camphor tree molecular marker technology, and more specifically relates to a camphor tree whole-genome SSR molecular marker and its preparation method and its application in the analysis of ancient camphor genetic diversity and identification of kinship. Background technique: [0002] Microsatellite (Microsatellite) is also called short tandem repeats (Short Tandem Repeats, STRs), simple sequence repeats (Simple Sequence Repeat, SSRs), simple sequence length polymorphism (SSLP). It refers to a DNA sequence that is repeated in tandem multiple times in units of 1 to 6 nucleotides. It is commonly found in the genomes of eukaryotes and prokaryotes, and even in virus genomes. SSR molecular markers have the following characteristics: repeat units and repeat numbers are highly variable, polymorphic information capacity is high; co-dominant, following Mende...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Patents(China)
IPC IPC(8): C12Q1/6895C12Q1/6806C12N15/11
CPCC12Q1/6806C12Q1/6895C12Q2600/156C12Q2531/113C12Q2525/151
Inventor 伍艳芳徐海宁王建汪信东郑永杰刘新亮戴小英邱凤英杨海宽
Owner JIANGXI ACAD OF FORESTRY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com