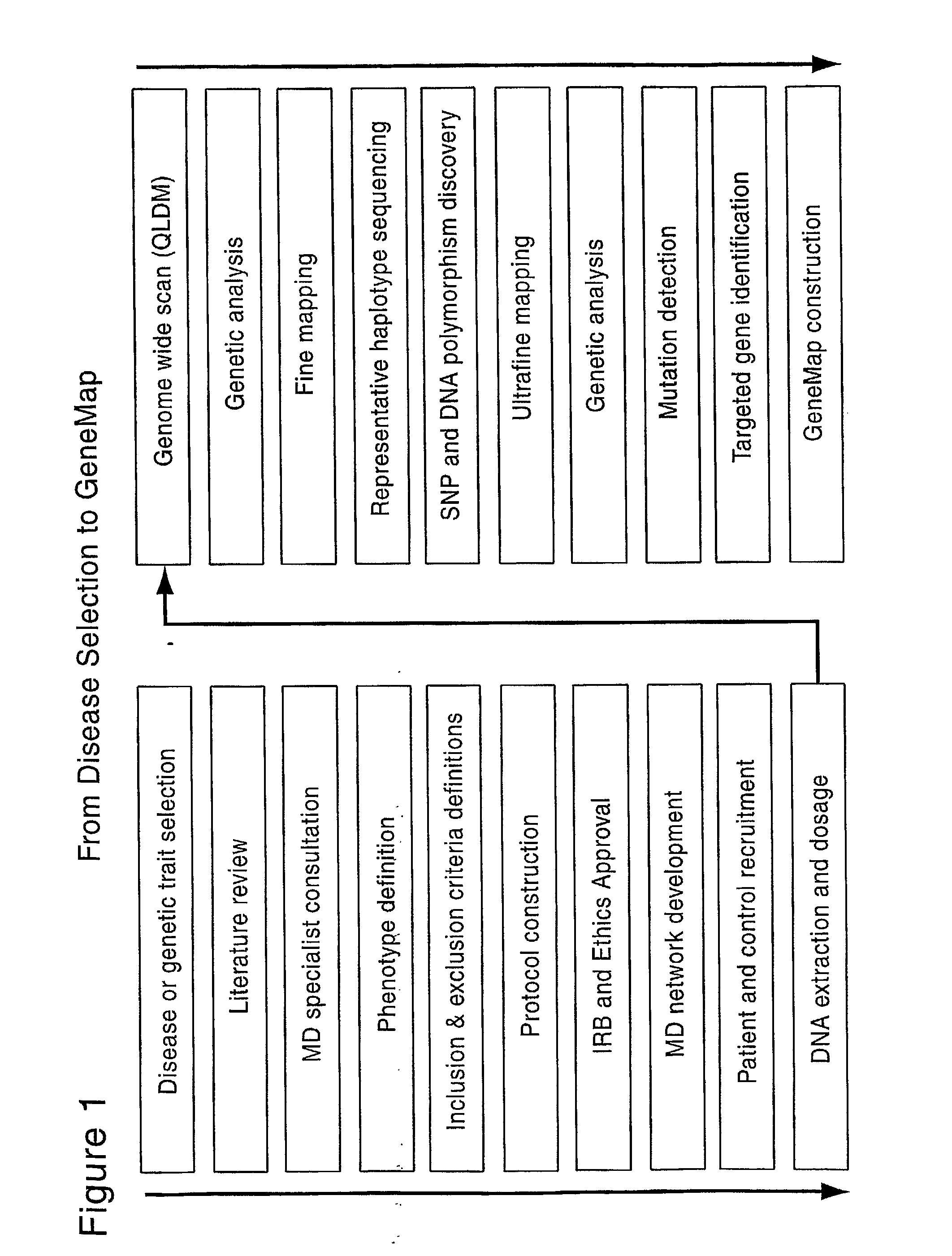
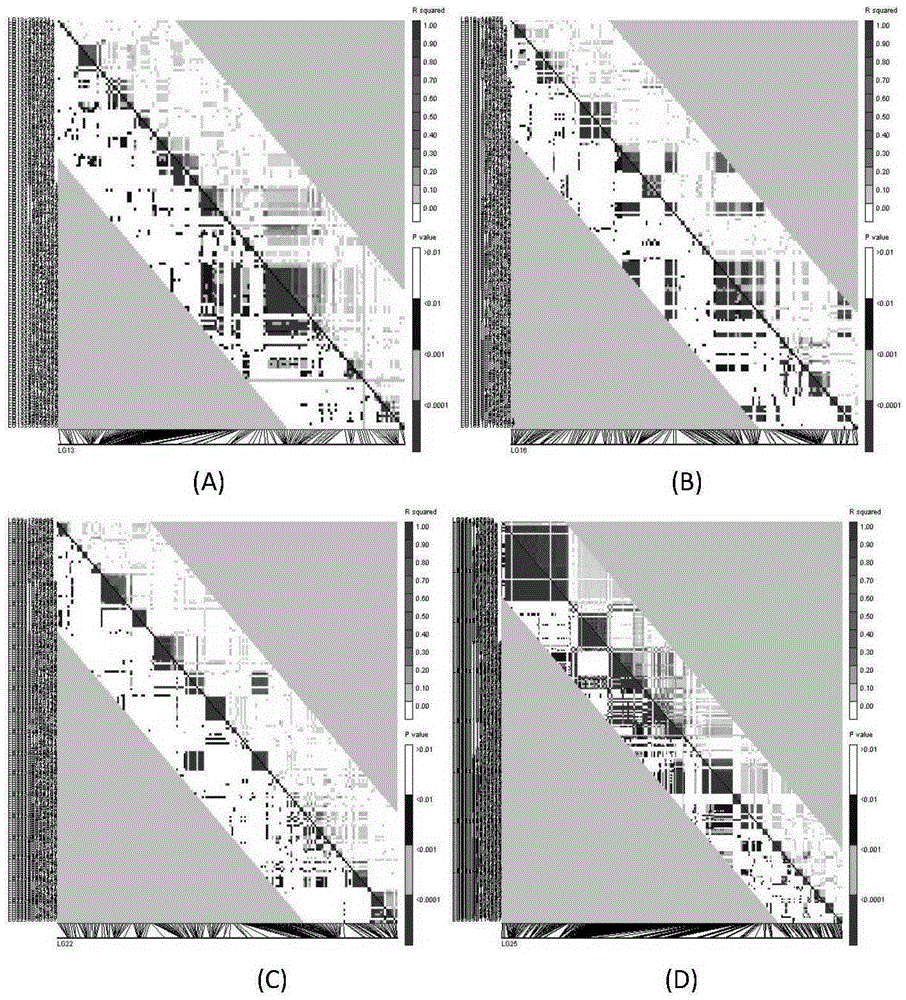
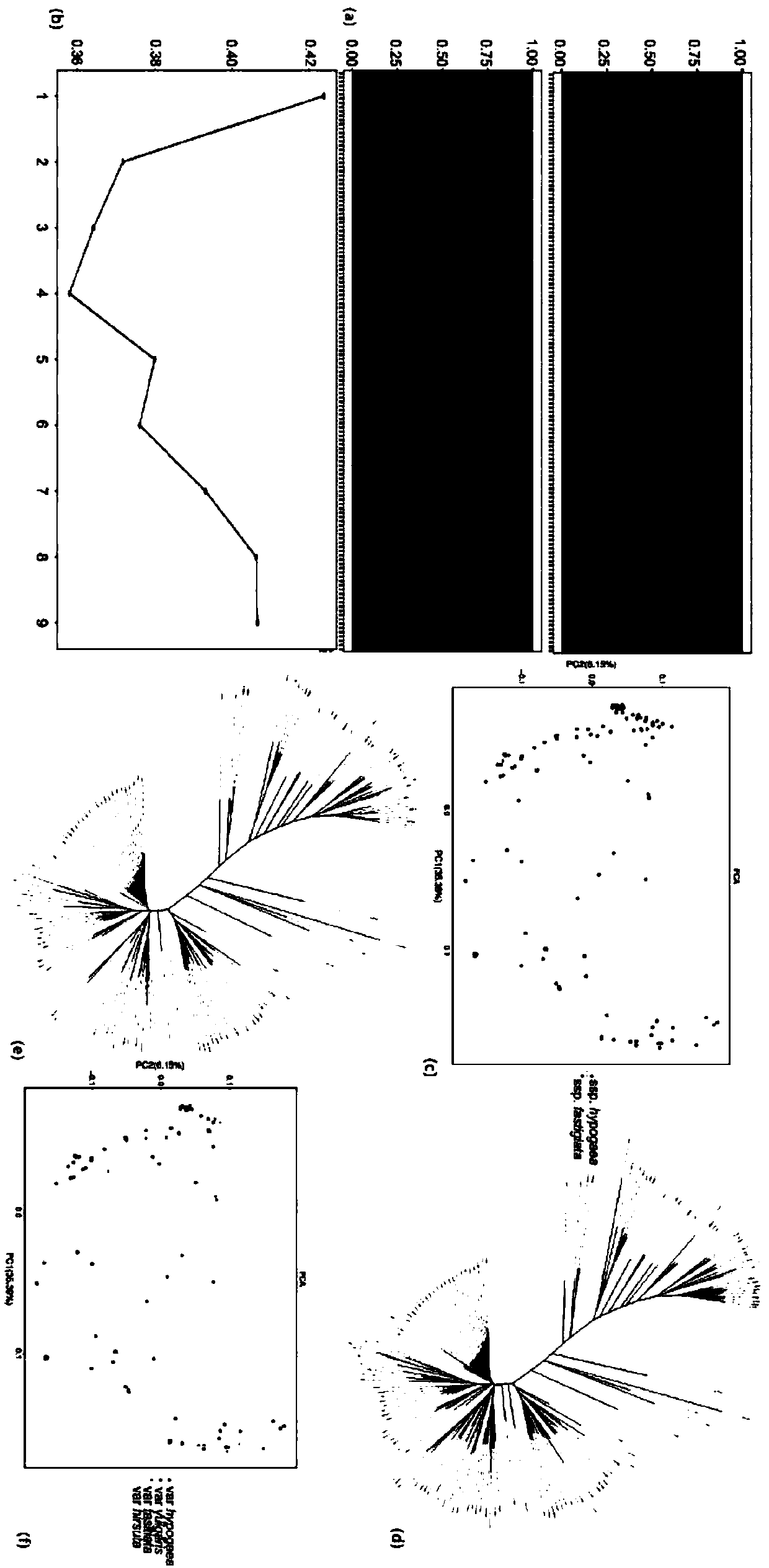
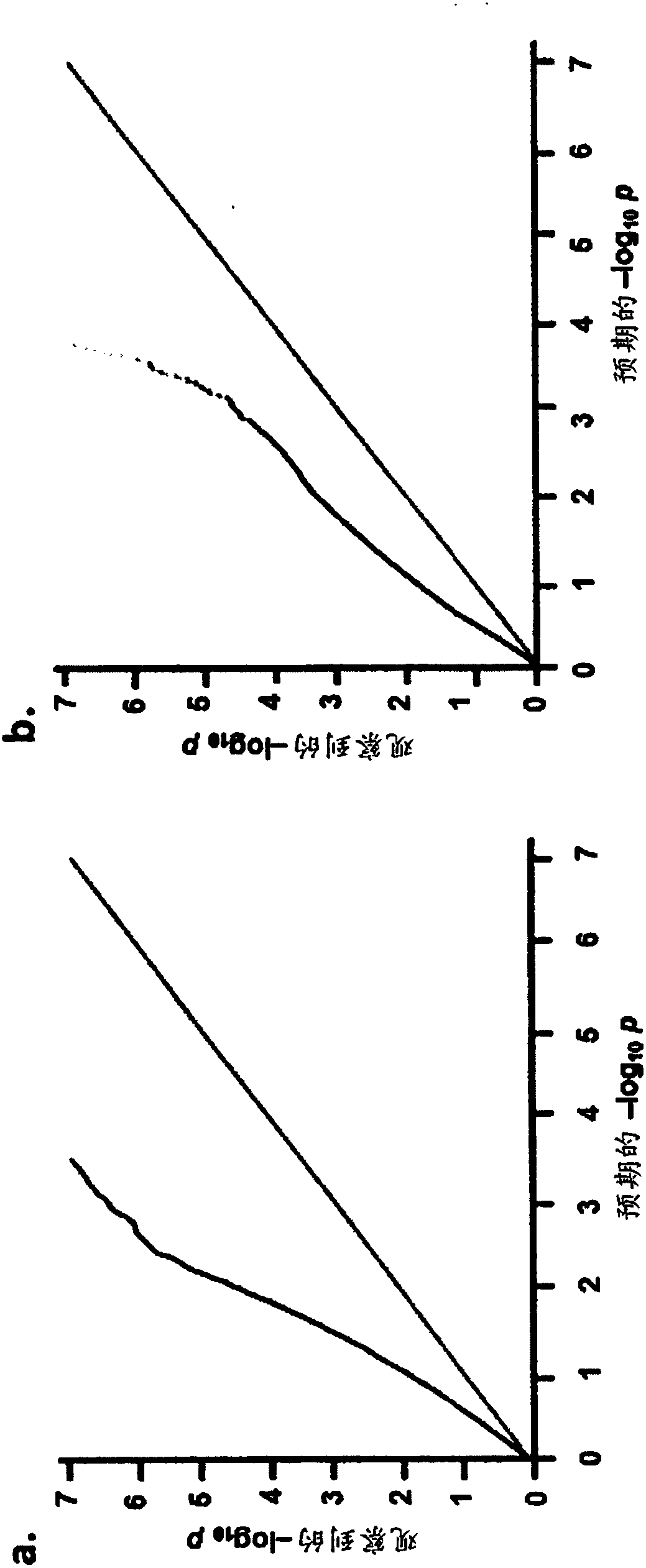
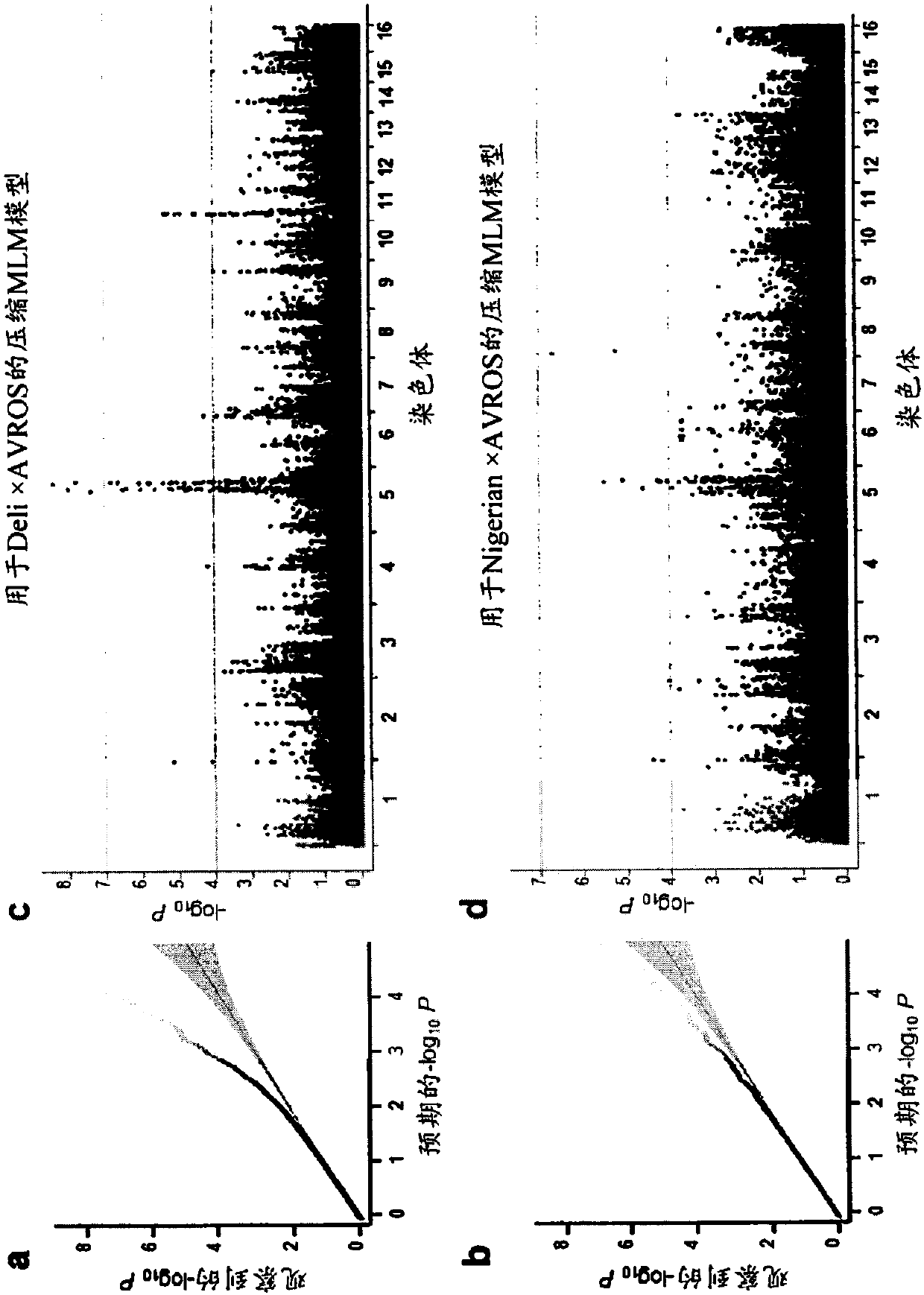
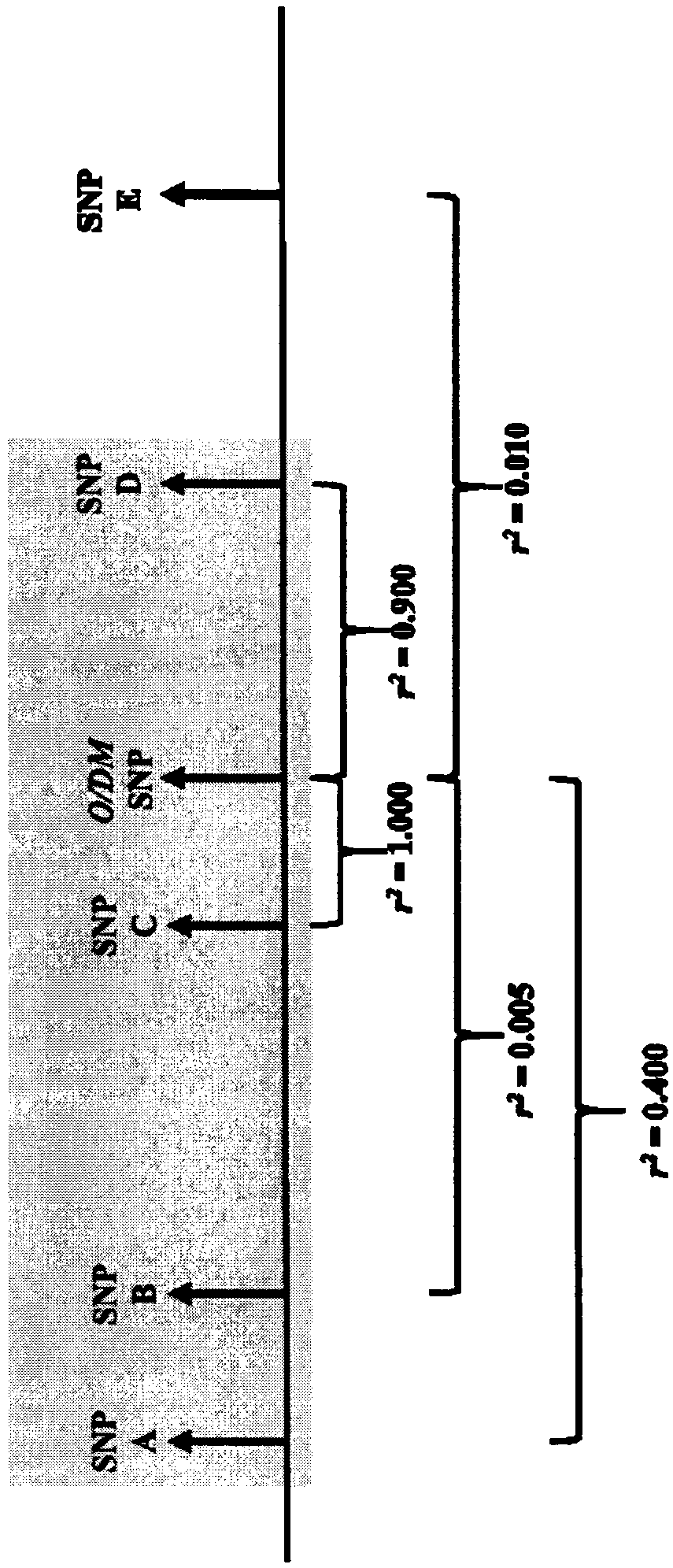
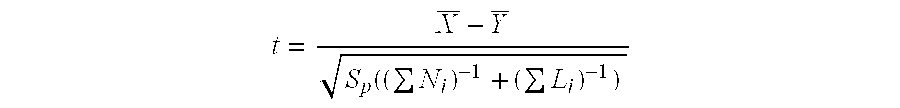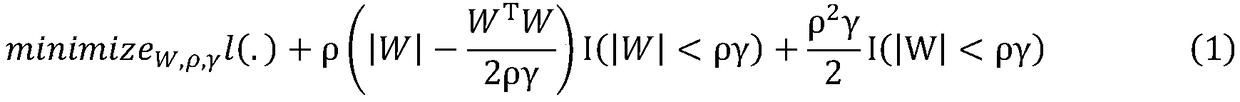Patents
Literature
69 results about "Linkage disequilibrium" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
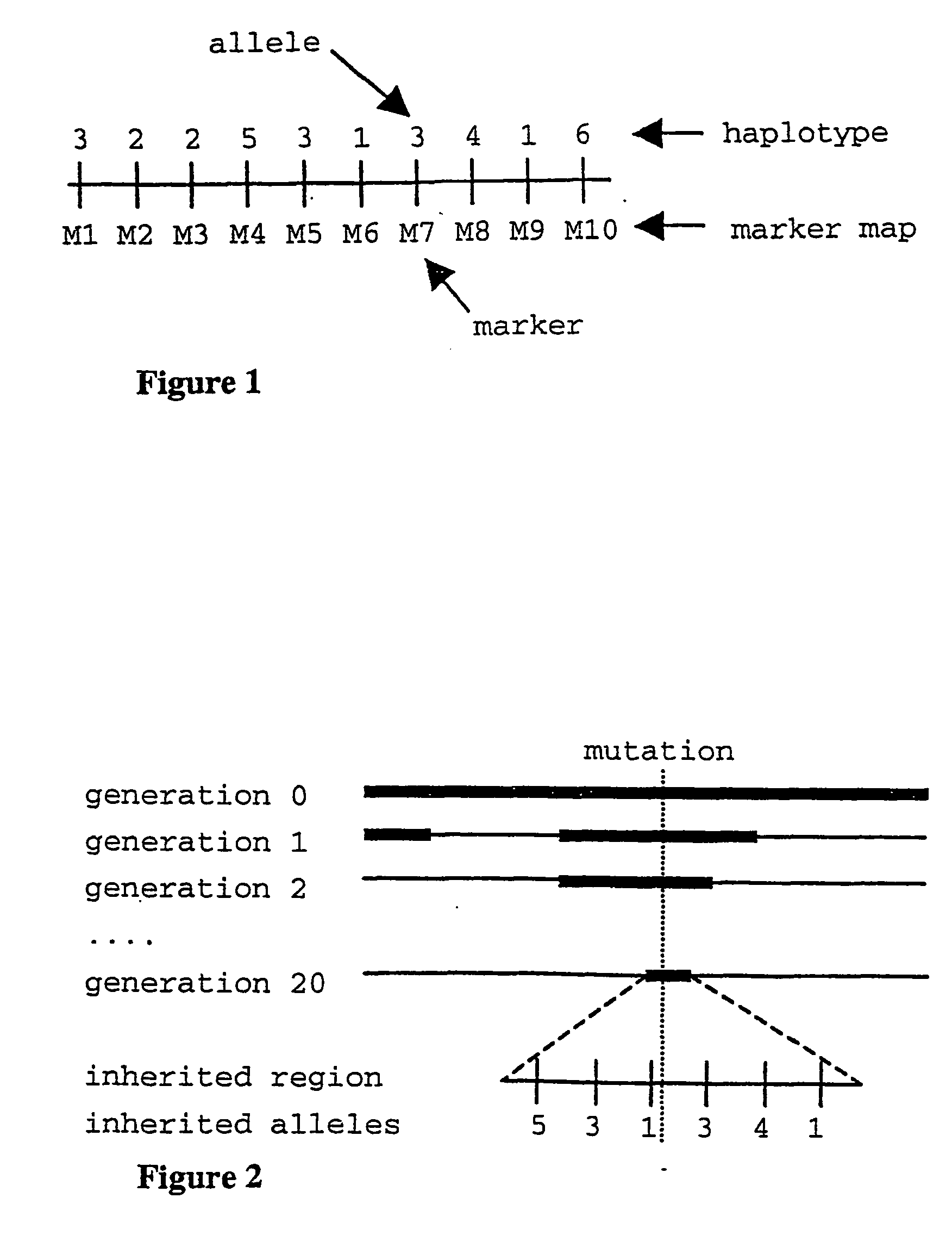
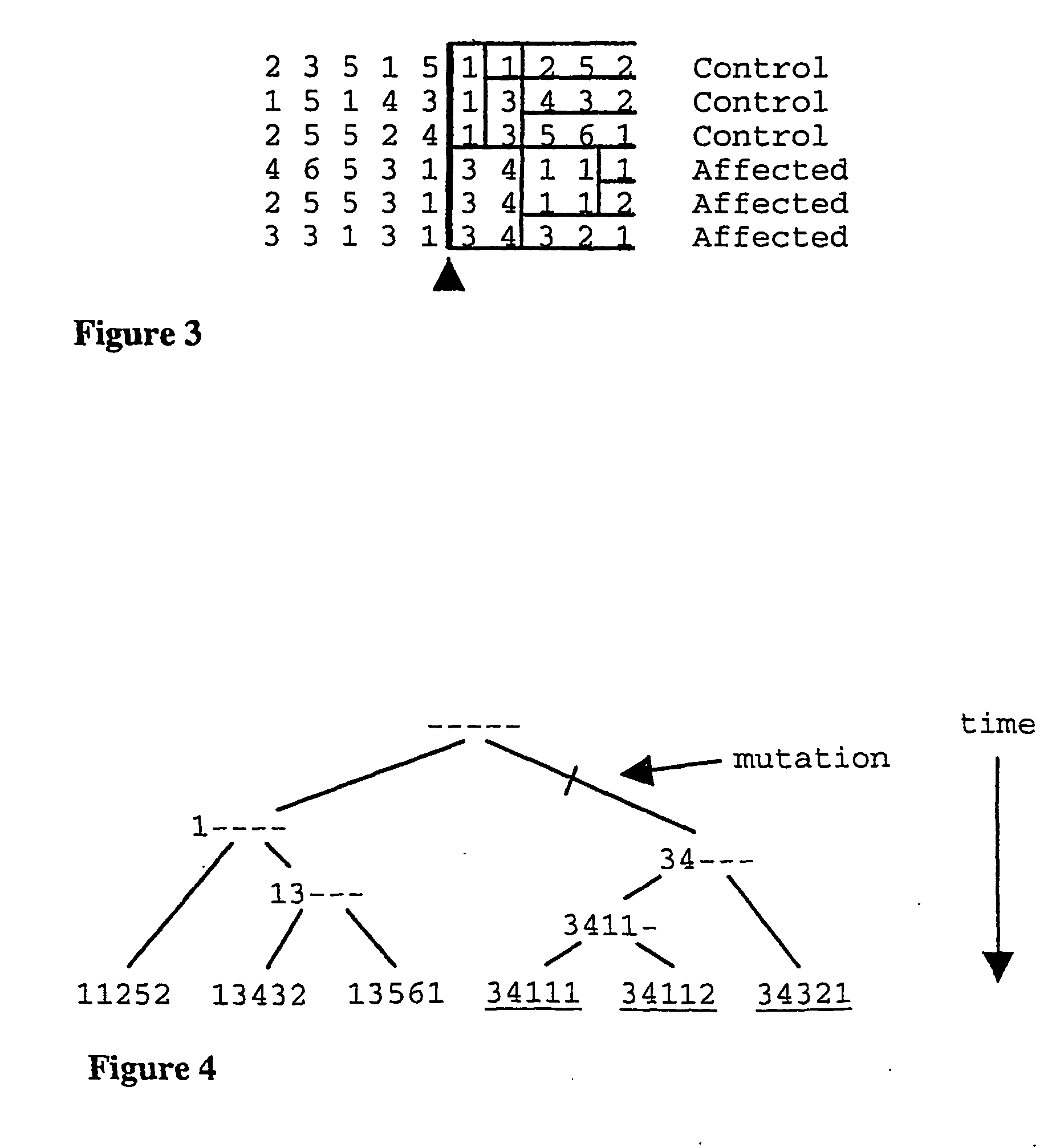
In population genetics, linkage disequilibrium is the non-random association of alleles at different loci in a given population. Loci are said to be in linkage disequilibrium when the frequency of association of their different alleles is higher or lower than what would be expected if the loci were independent and associated randomly.
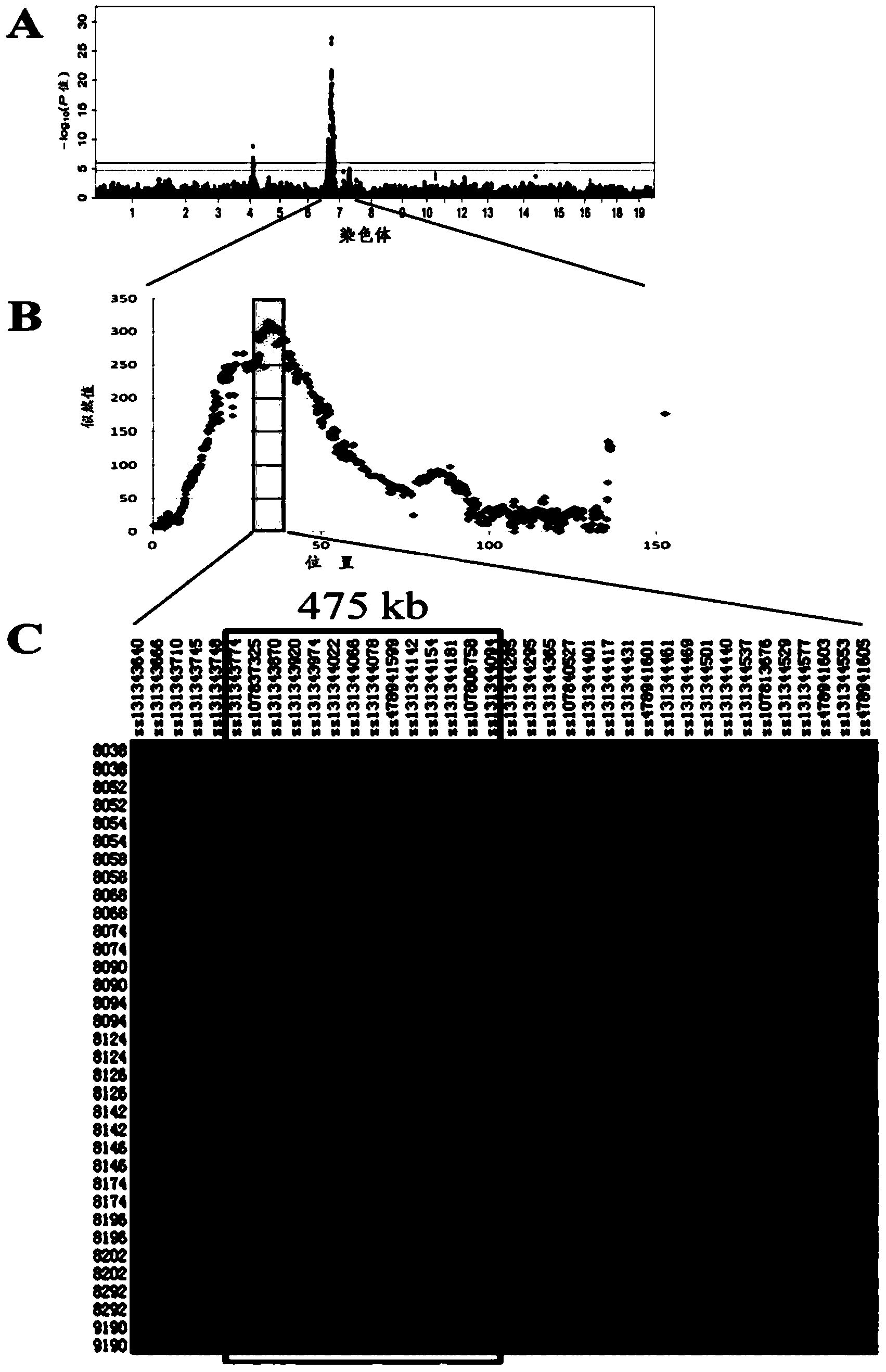
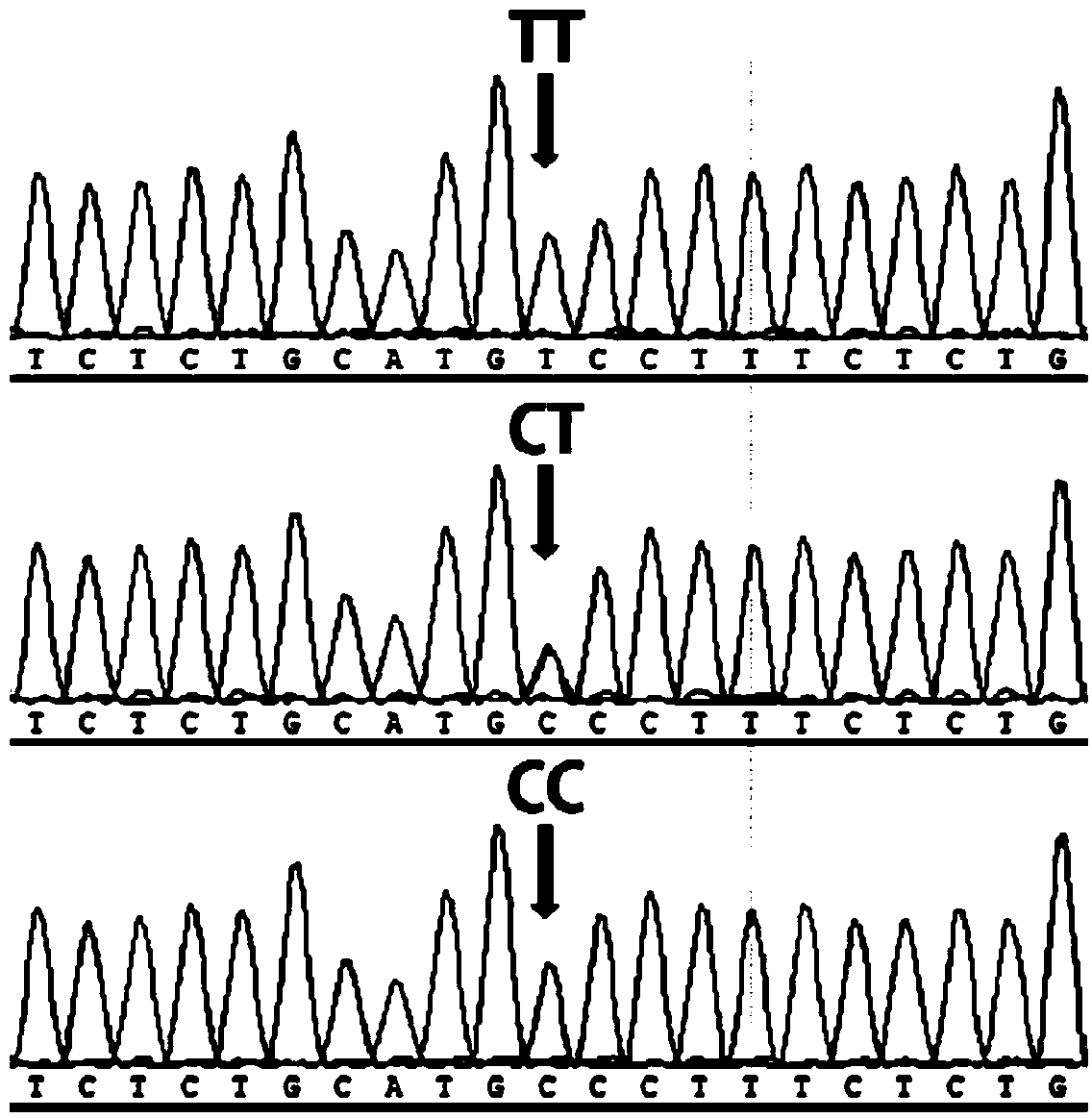
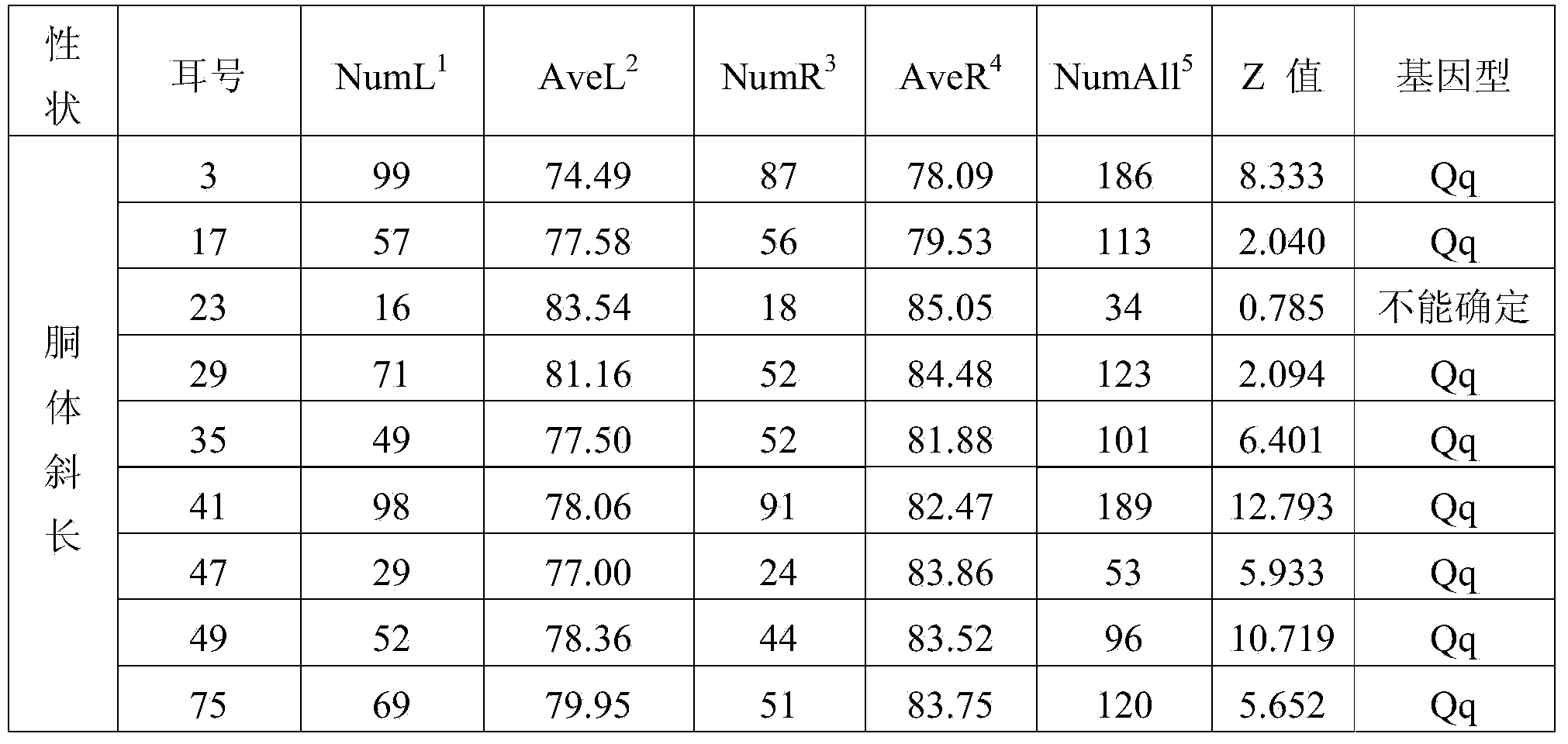
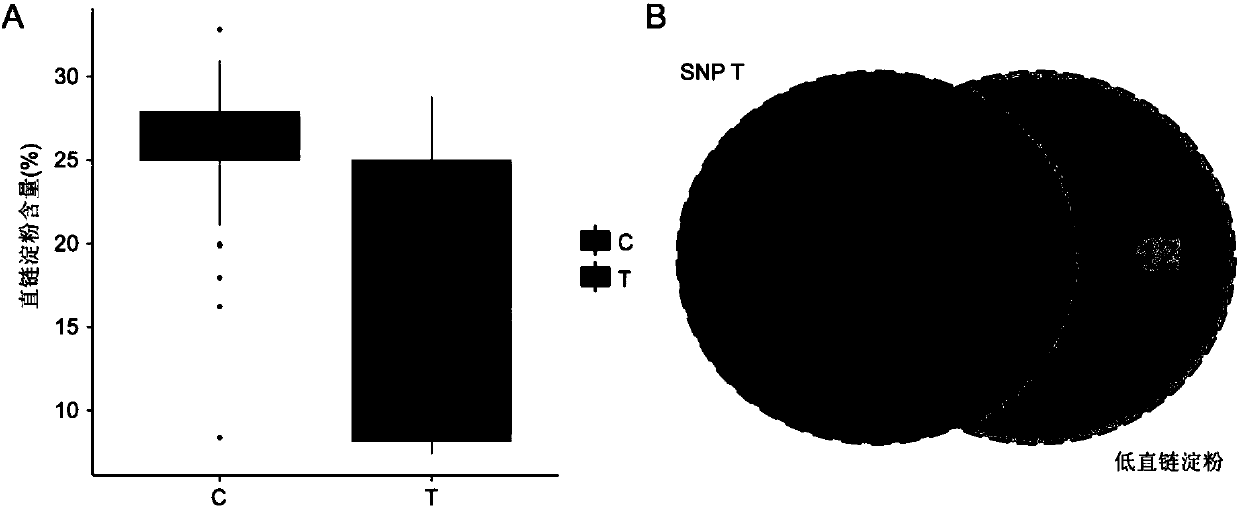
Major SNP (single nucleotide polymorphism) marker influencing growth traits of pigs and application thereof in genetic improvement of productivity of breeding pigs
The invention provides a major SNP (single nucleotide polymorphism) marker influencing the growth traits of pigs. The SNP marker is located at a nucleotide sequence of an HMGA1 gene of a pig chromosome 7, a locus of the SNP marker is the nucleotide mutation of C857-G857 with an SEQ ID NO:1 sequence labeling position of 857, corresponding to a 34983991st nucleotide locus C> on chromosome 7 in a reference sequence in an international pig genome version 10.2, G mutation, or one of seven other loci completely linked with the locus. The invention also provides the application of the SNP marker in the genetic improvement of growth traits of breeding pigs, and provides the application of an SNP molecular marker with a linkage disequilibrium degree (r2) with the SNP marker of greater than 0.8 in the genetic improvement of growth traits of breeding pigs. The growth traits include one or more of the length and height of a living body of a pig, carcass length, carcass weight, daily gain and head weight. According to the invention, the breeding process of breeding pigs can be accelerated, the productivity of breeding pigs can be effectively improved, and remarkable economic benefits can be obtained.
Owner:JIANGXI AGRICULTURAL UNIVERSITY
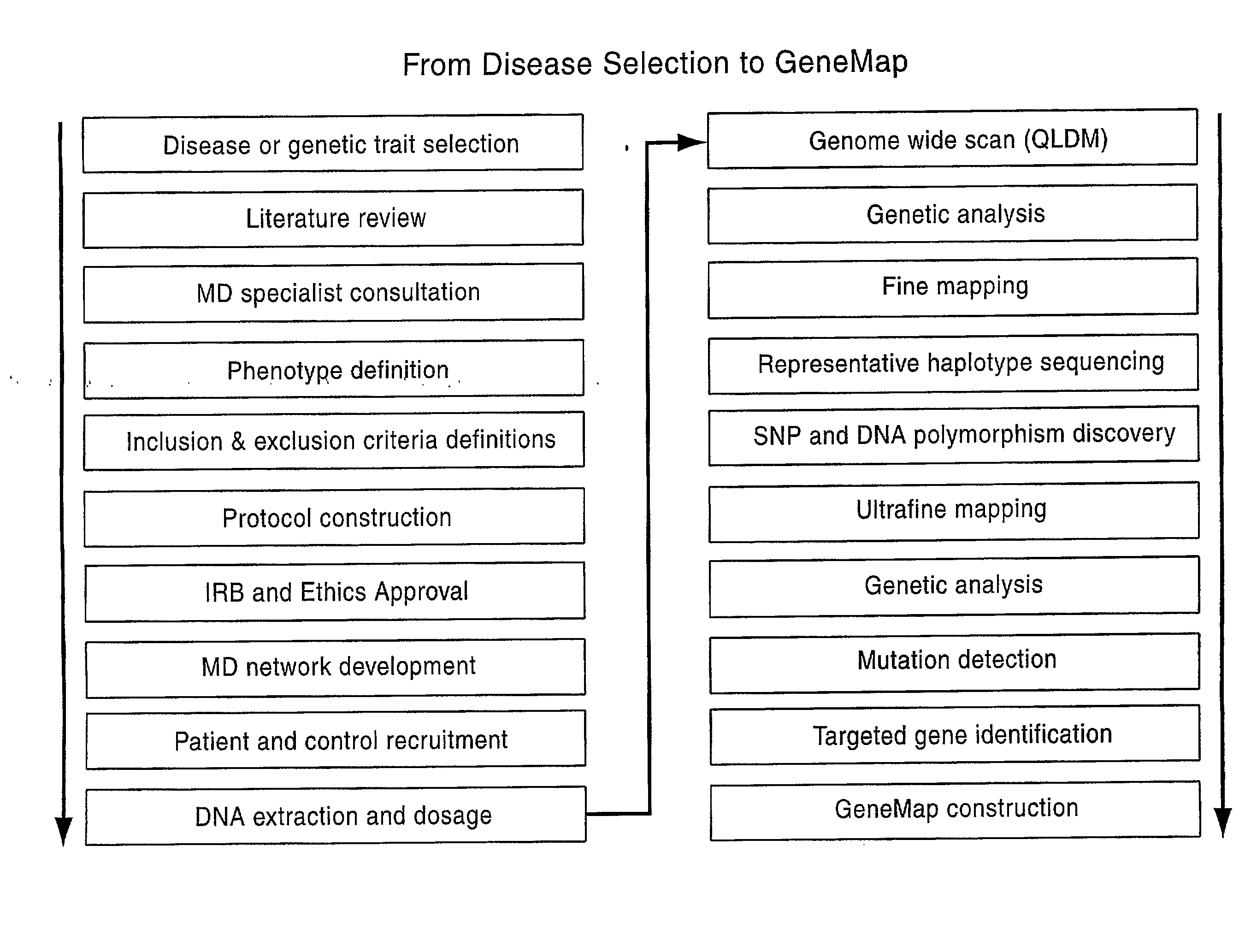
Genemap of the human genes associated with longevity
InactiveUS20090305900A1Electrolysis componentsVolume/mass flow measurementGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of SNP markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's longevity, their protection against age-related diseases and / or their response to a particular drug or drugs.
Owner:BELOUCHI ABDELMAJID +12
Genemap of the human genes associated with crohn's disease
InactiveUS20090081658A1Reduce frequencyTreatment safetyMicrobiological testing/measurementGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of polymorphism markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to Crohn's disease and / or their response to a particular drug or drugs.
Owner:GENIZON BIOSCI
Upland cotton SNP marker and application thereof
InactiveCN105349537AImprove detection efficiencyImprove work efficiencyNucleotide librariesMicrobiological testing/measurementAgricultural scienceHomologous sequence
The invention discloses an upland cotton SNP marker and application thereof, and belongs to the field of upland cotton SNP marker development. The upland cotton SNP marker is obtained by developing a single-copy SNP marker of an upland cotton genome via chip hybridization and homologous sequence comparison, and picking out SNP markers in a linkage disequilibrium recession distance via linkage disequilibrium analysis. The SNP marker provided by the invention can be applied to analysis of genetic diversity and group structure, or to germplasm identification of upland cotton, is single-copy, and has no remarkable linkage disequilibrium among markers, so that the efficiency of detection, adopting the SNP marker, during analysis of genetic diversity, group structure or fingerprint identification can be greatly improved, and the work efficiency is higher.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Genemap of the human genes associated with psoriasis
The present invention relates to the selection of a set of polymorphism markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to psoriasis disease and / or their response to a particular drug or drugs.
Owner:GENIZON BIOSCI
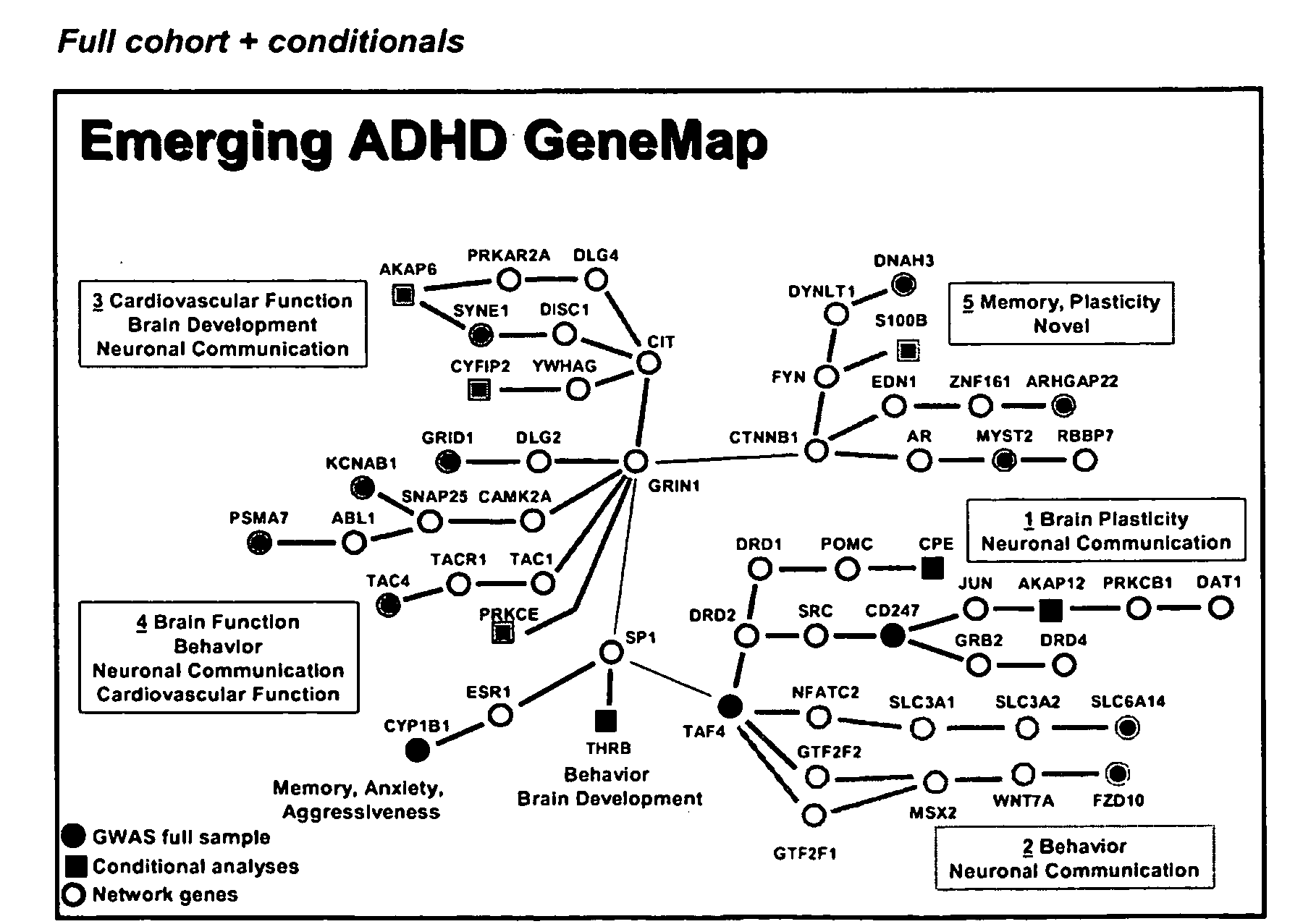
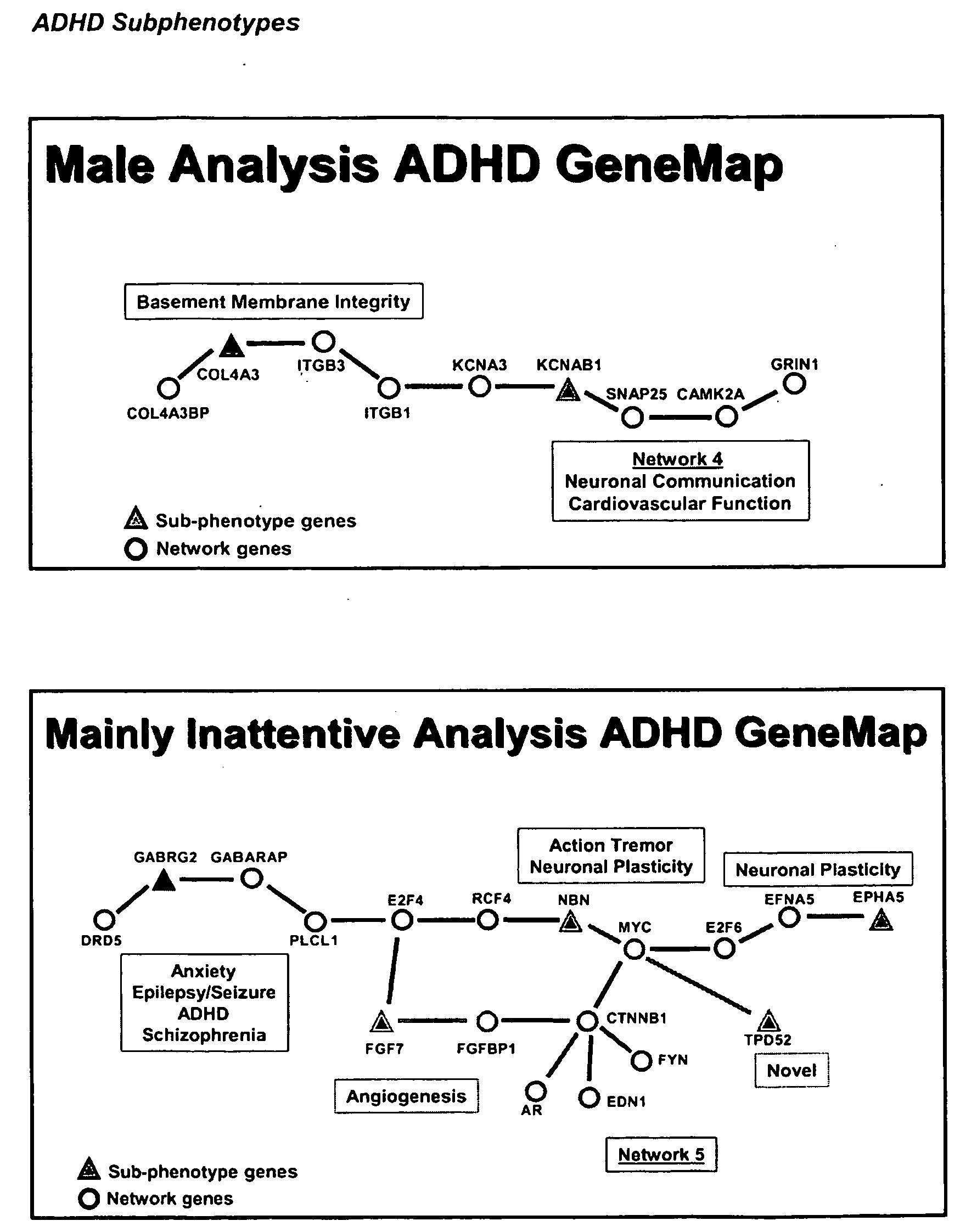
Genemap of the human genes associated with adhd
InactiveUS20100120628A1Easy to understandConvenient treatmentMicrobiological testing/measurementLibrary screeningGenomicsDisease
The present invention relates to the selection of a set of polymorphism markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to ADHD disease and / or their response to a particular drug or drugs.
Owner:GENIZON BIOSCI
Genemap of the human genes associated with crohn's disease
InactiveUS20090181380A1Microbiological testing/measurementAnalogue computers for chemical processesGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of polymorphism makers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to IBD (ex: Chrohn's disease) and / or their response to a particular drug or drugs.
Owner:BELOUCHI ABDELMAJID +12
Chinese population linkage analysis single nucleotide polymorphism (SNP) marker sets and use method and application thereof
InactiveCN102121046AEnsure reliabilityEnsure comprehensivenessMicrobiological testing/measurementDNA/RNA fragmentationGenetic linkage disequilibriumInternational HapMap Project
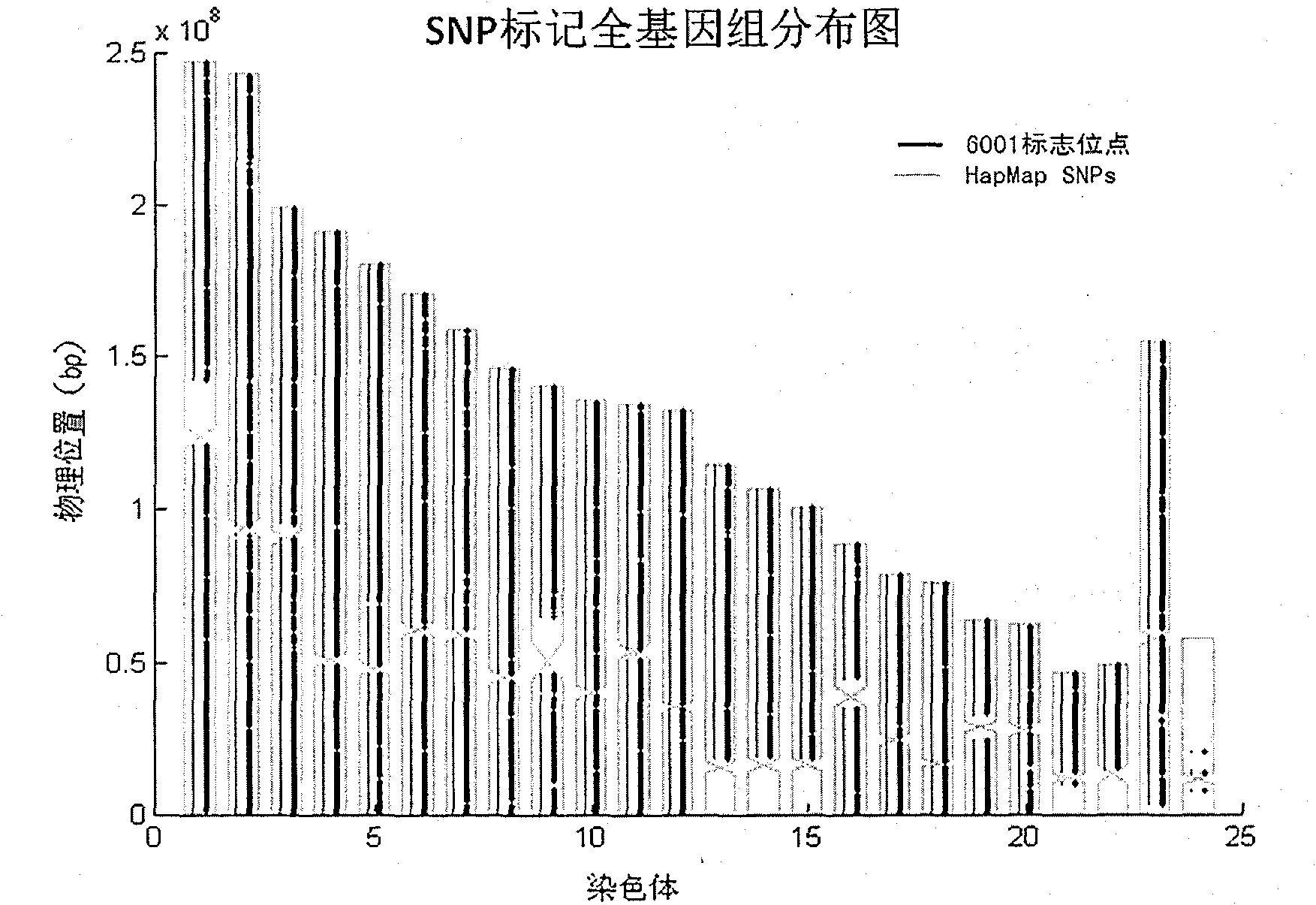
The invention relates to Chinese population linkage analysis single nucleotide polymorphism (SNP) marker sets and a use method and application thereof. On the basis of hundreds of millions of Chinese Han population data results in the mass data of the International HapMap Project, medium-density and high-density SNP marker sets for linkage analysis are constructed and optimized, according to the statistical comparisons of multiple parameters such as the linkage disequilibrium, the polymorphism level, the typing success rate, the distribution position and density of genomes and the functional characteristic, and the multi-level selections and experimental verifications. The two marker sets separately contain 3000 and 6001 loci, wherein the 6001 loci contain the 3000 loci. The SNP sets aim at the Han genetic background in design, have high polymorphism in Chinese and can realize the aim of efficiently marking the Chinese family sample genomes. The selection of polymorphic loci is based on the neutral evolution principle, and all the loci are in a non-gene function region, thus the influence of evolution on the gene function can be avoided. Meanwhile, the characteristics that the marking loci have high typing detectability and can uniformly cover the whole genomes can ensure that the whole genomes can be screened completely and new pathogenic genes can be located and found. The two sets of SNP markers are used to customize probes or chips and perform whole-genome genotyping to family samples; and the typing data are used for linkage analysis, and the haplotyping and fine locating of the linkage candidate region are also adopted, thus the use method has more accurate locating result than the traditional method while the cost is lower and the speed is higher. The distribution and coverage of the 6001 SNP marker set in human chromosomes are shown in the appended drawings.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION +1
Method for prediction of human iris color
InactiveUS20110312534A1Sugar derivativesMicrobiological testing/measurementGenotypingLinkage disequilibrium
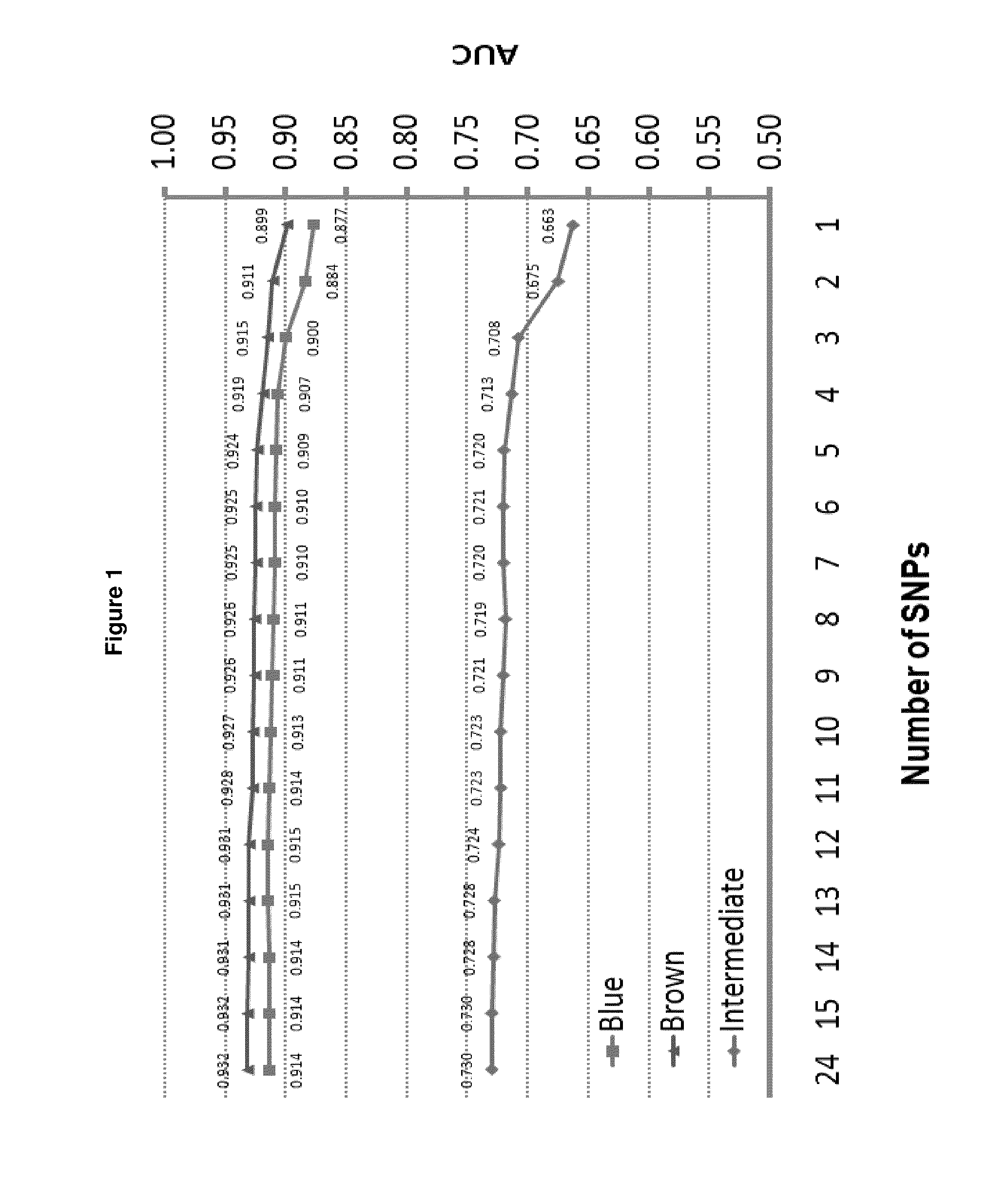
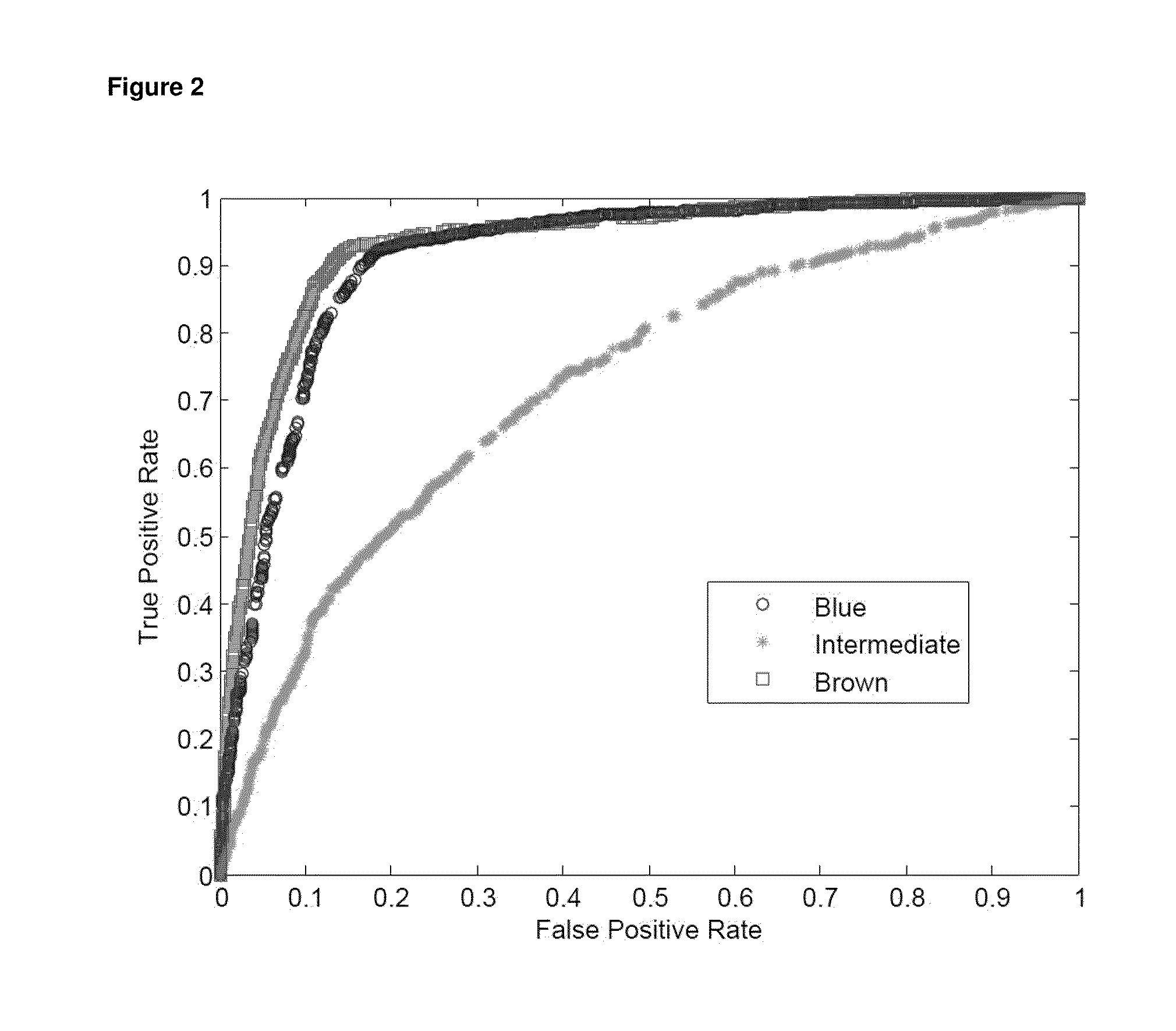
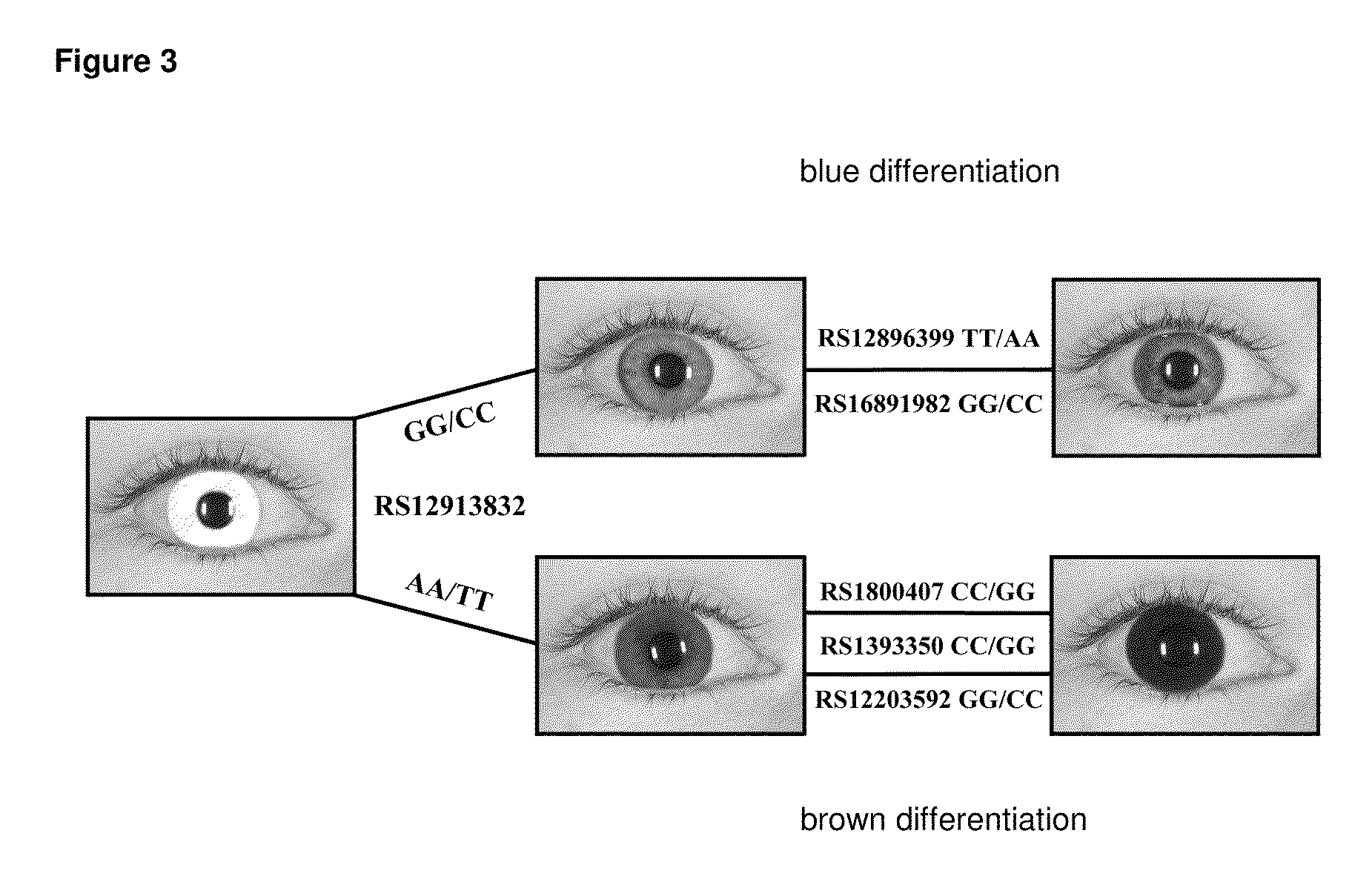
A method for predicting the iris color of a human, the method comprising:(a) obtaining a sample of the nucleic acid of the human;(b) genotyping the nucleic acid for at least the following polymorphisms:(i) the single nucleotide polymorphism (SNP) rs12913832 or a polymorphic site which is in linkage disequilibrium with rs12913832 at an r2 value of at least 0.9;(ii) the SNP rs1800407 or a polymorphic site which is in linkage disequilibrium with rs1800407 at an r2 value of at least 0.5; and,(iii) the SNP rs12896399 or a polymorphic site which is in linkage disequilibrium with rs12896399 at an r2 value of at least 0.5; and(c) predicting the iris color based on the results of step (b).A method of genotyping said polymorphisms, and kits comprising or a solid substrate having attached thereto nucleic acid molecules suitable for performing the method.
Owner:ERASMUS UNIV MEDICAL CENT ROTTERDAM ERASMUS MC
Markers and methods for assessing and treating severe or persistant asthma and TNF related disorders
InactiveUS20100331209A1Decreased asthma exacerbationPredict suitabilityNucleotide librariesMicrobiological testing/measurementMolecular Targeted TherapiesLinkage disequilibrium
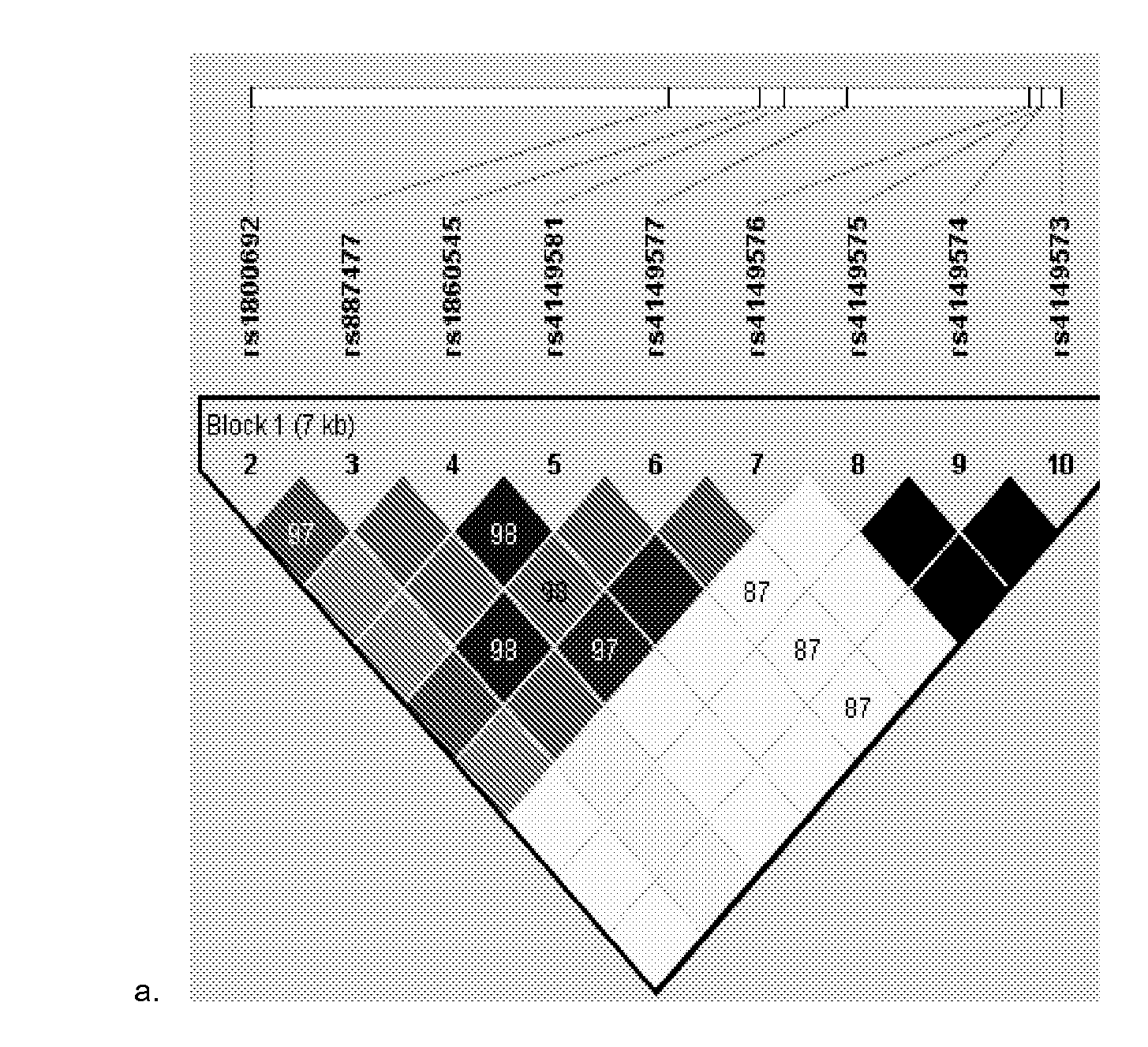
A method for assessment of the suitability of and / or effectiveness of a target therapy for a TNF-mediated-related disorder, such as severe or persistent asthma, in a subject evaluates the presence, absence, and / or magnitude of expression of one or more genes corresponding to contacting the sample with a panel of nucleic acid segments consisting of at least a portion of at least one member from the group consisting of the nucleotide sequences corresponding to at least one of TNFRSF1A SNP rs4149581 (SEQ ID NO:1), TNFRSF1 B SNP rs3766730 (SEQ ID NO:2) or TNFRSFI B SNP rs590977 (SEQ ID NO:3) SNPs which results in a determination that one or more of said SNPs in a sample are in linkage disequilibrium (LD). The method enables identification of the effectiveness of target therapies prior to or after starting a patient on such therapies.
Owner:CENTOCOR ORTHO BIOTECH
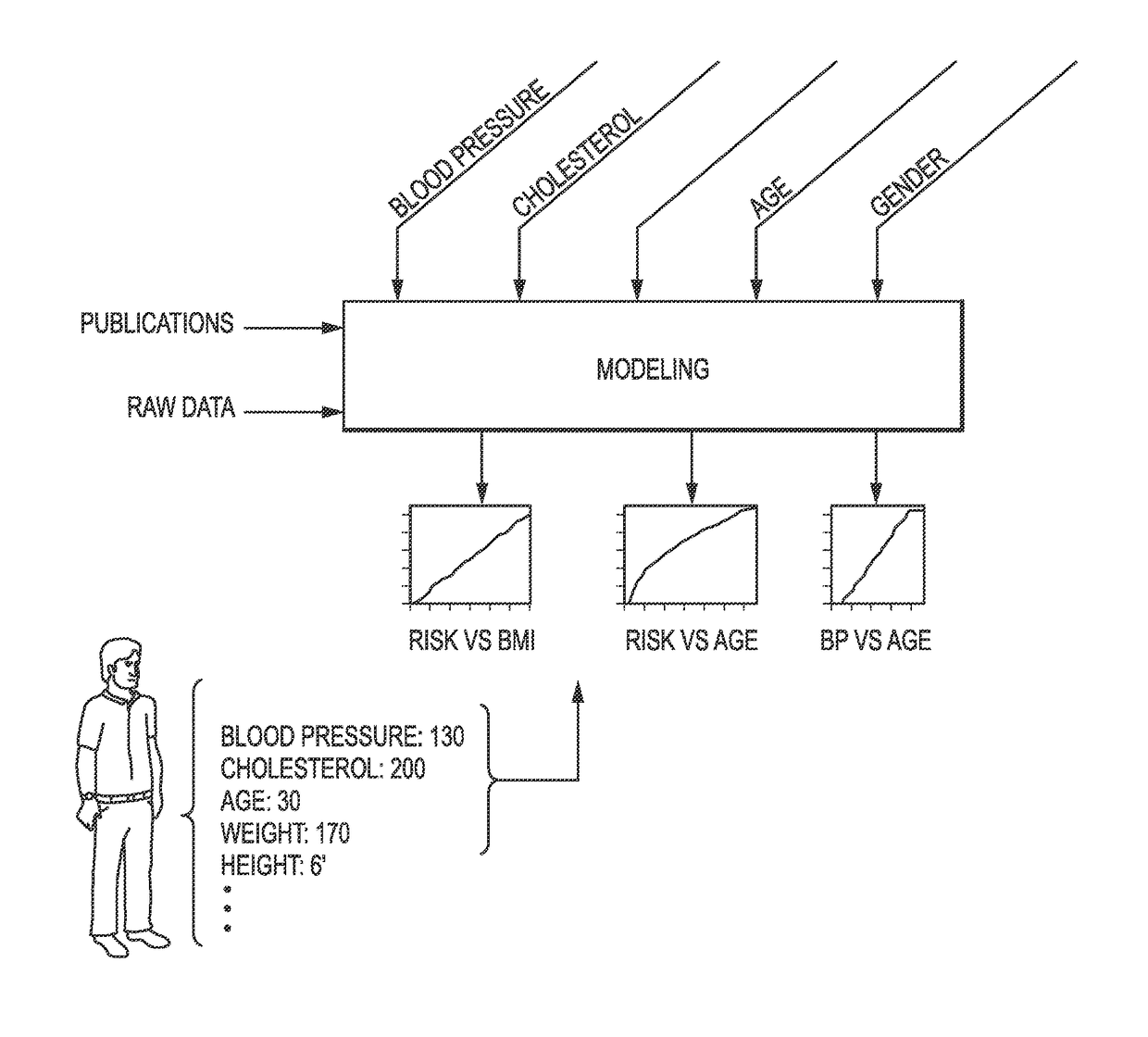
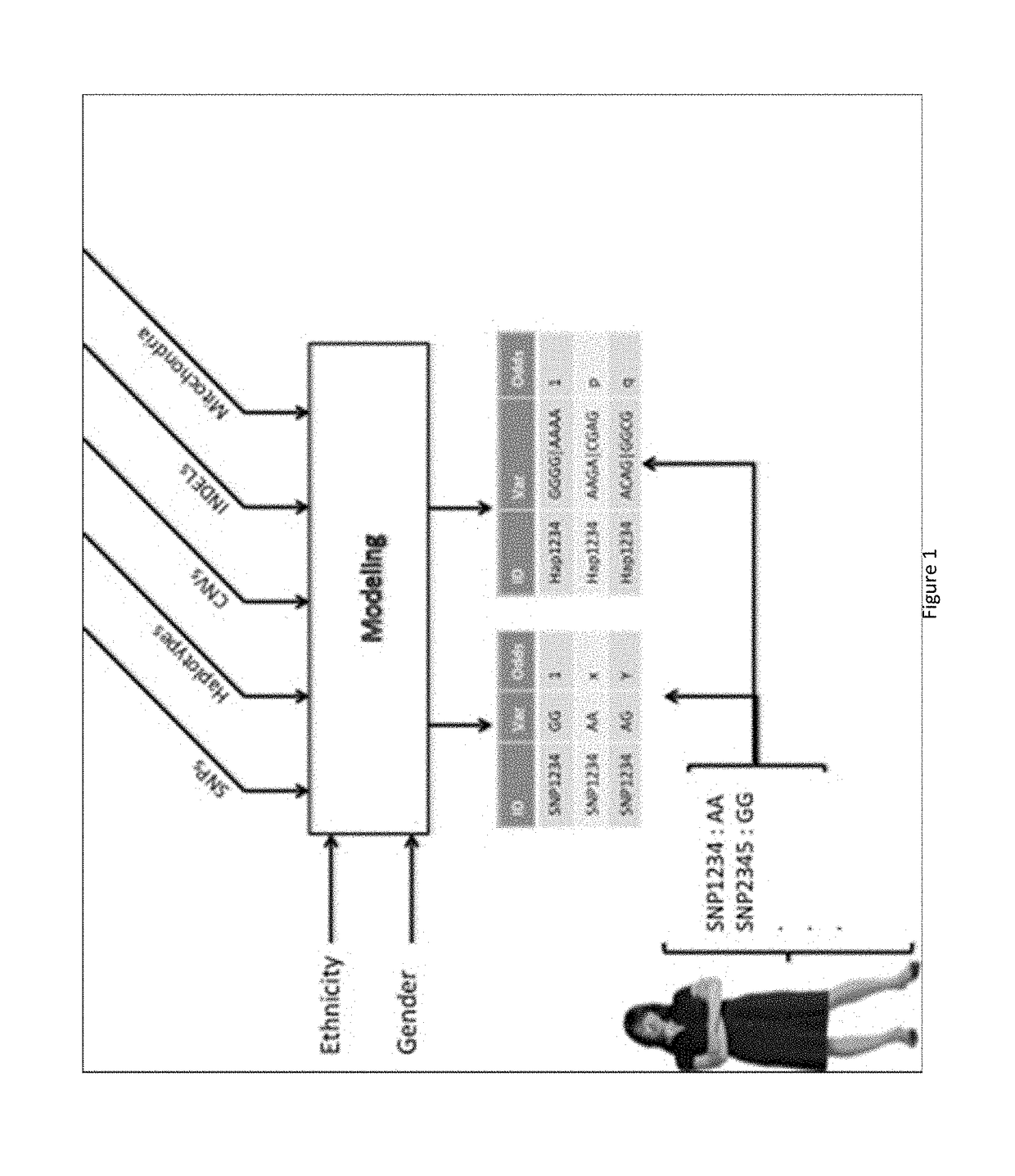
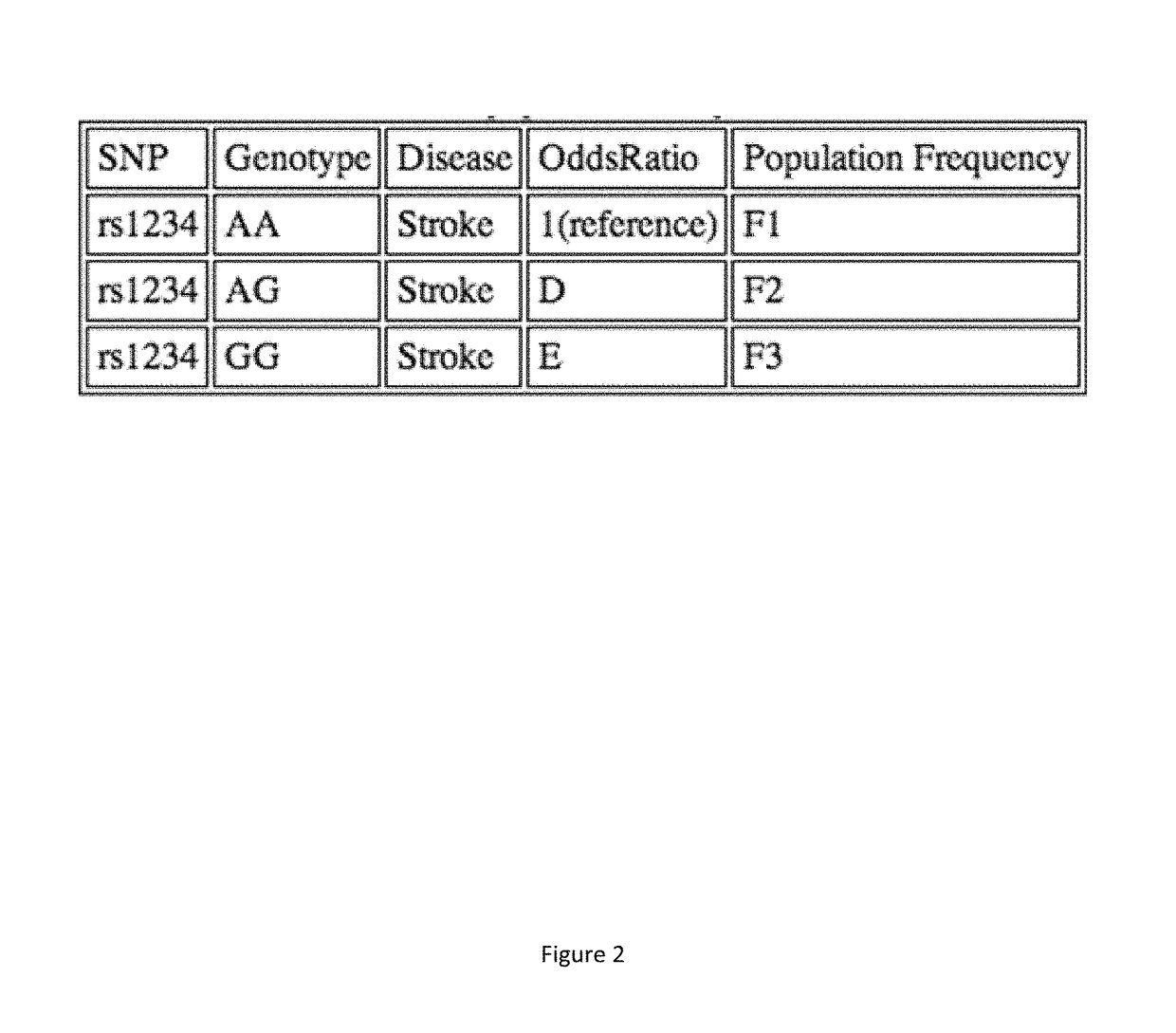
Genetic and environmental risk engine and methods thereof
Risk engines and related methods may benefit patients and their physicians. For example, patients may benefit from being able to determine their personal genetic, environmental, and behavioral risks. Moreover, physicians may be able to provide statistically-driven individual recommendations based on a risk engine's determination of such risks. A method can include selecting one or more candidate genetic variants associated with a phenotype from the scientific literature. The method can also include scoring a genetic association between the one or more candidate genetic variants and the phenotype. The method can further include selecting one or more high-scoring genetic variants. Selecting a best genetic variant within each of at least one linkage disequilibrium (LD) block can also be included in the method. The method can additionally include calculating risk associated with the best genetic variant from the at least one LD block.
Owner:RR HEALTH INC
Method for gene mapping from chromosome and phenotype data
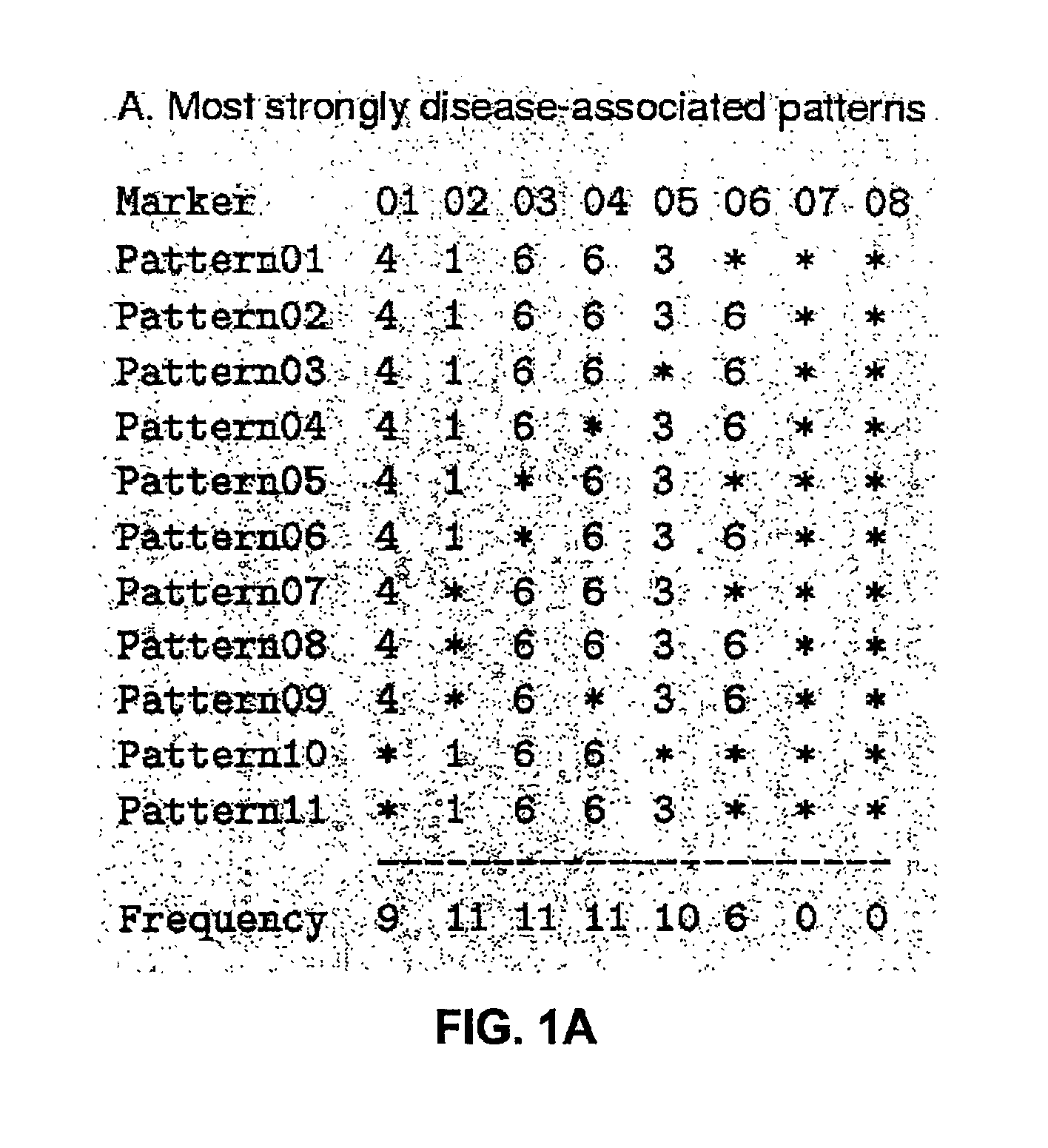
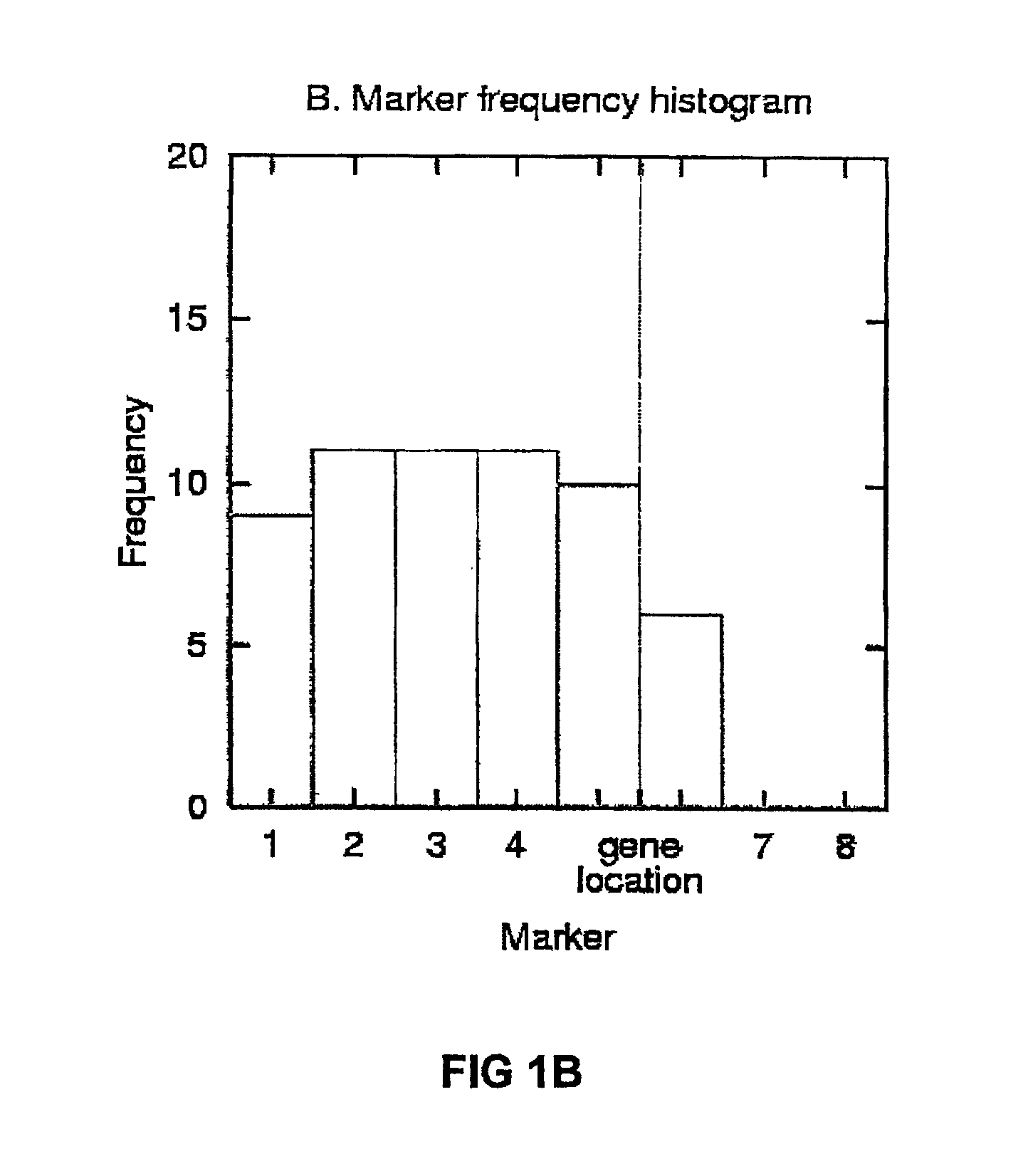
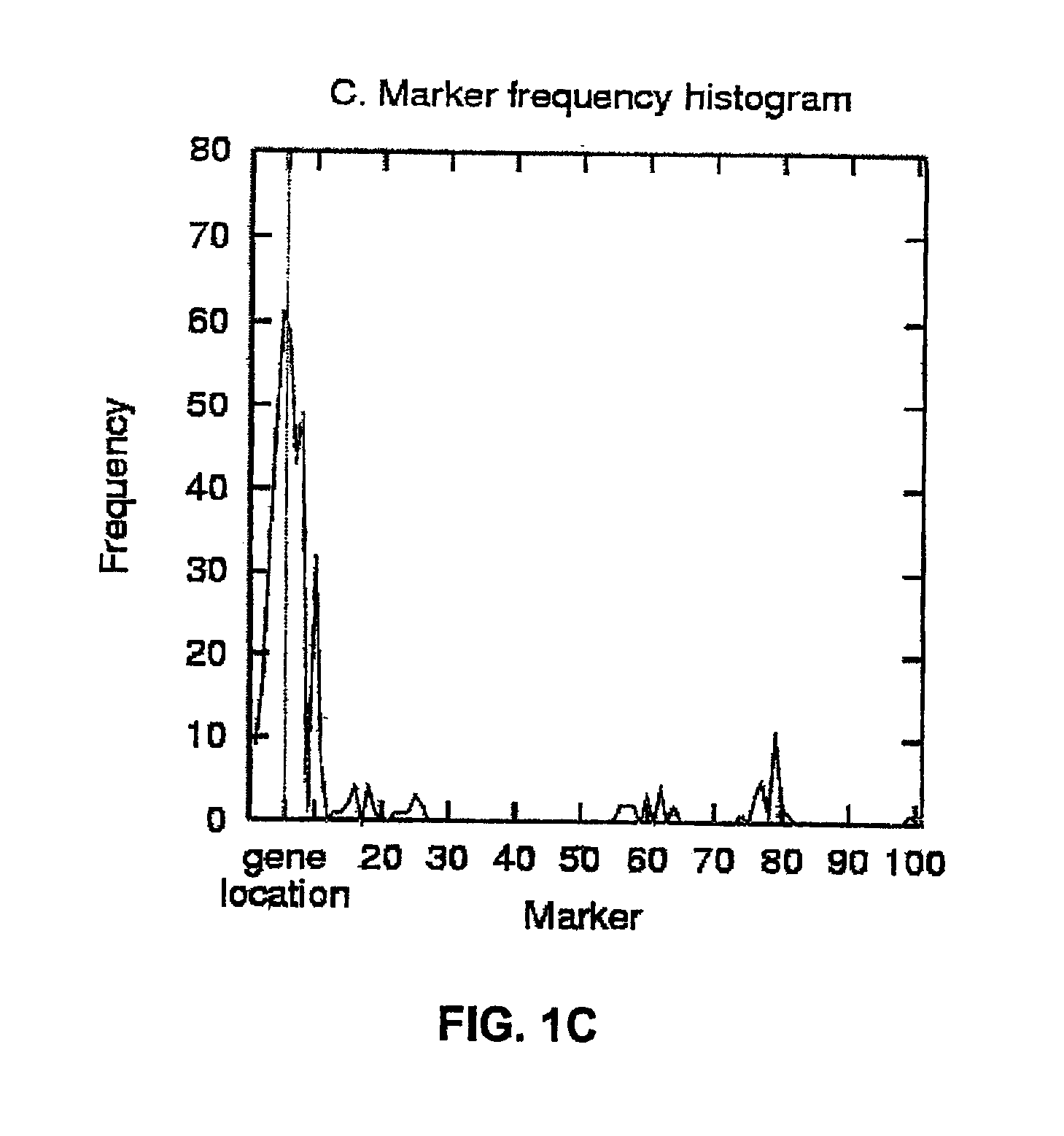
InactiveUS6909971B2Data processing applicationsMicrobiological testing/measurementGene mappingNucleotide
The present invention relates to a method for gene mapping from chromosome and phenotype data, which utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. All marker patterns P that satisfy a certain pattern evaluation function e(P) are searched from the data, each marker mi of the data is scored by a marker score and the location of the gene is predicted as a function of the scores s(mi) of all the markers mi in the data.
Owner:LICENTIA OY
Method for developing waxy1-gene internal molecular markers on basis of association analysis and KASP
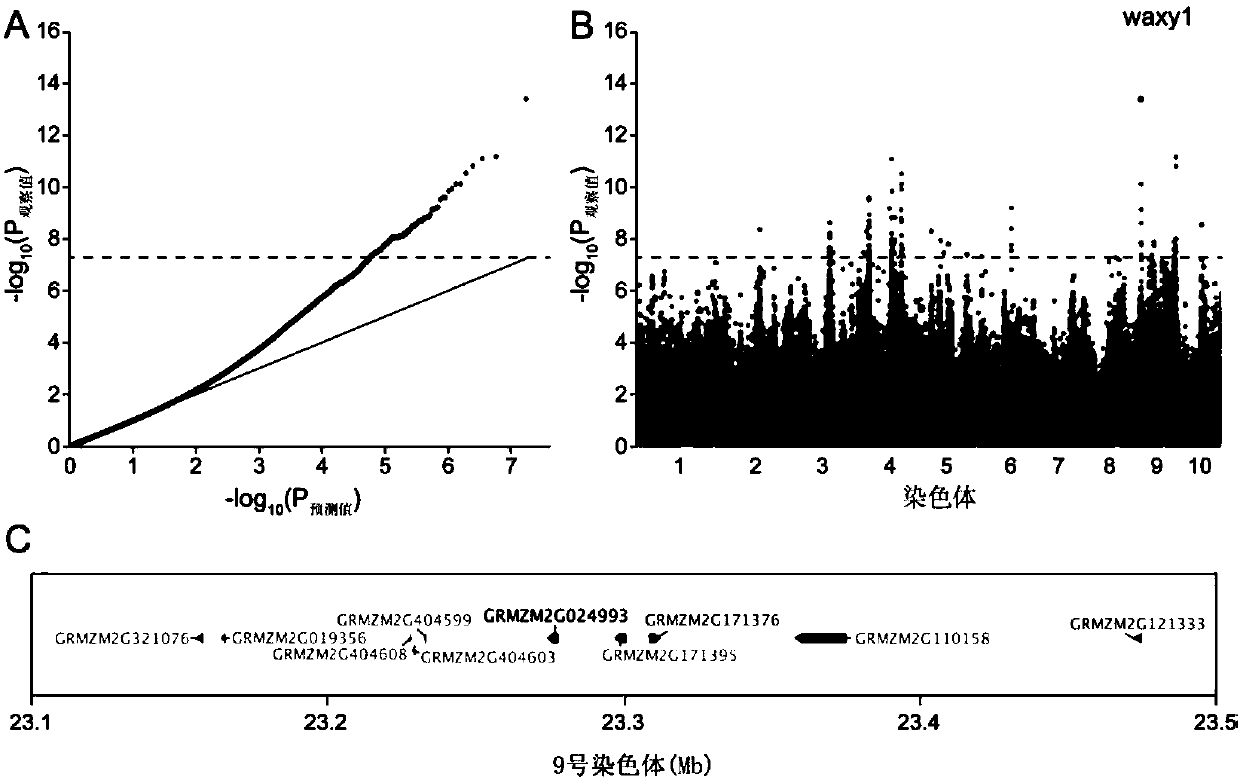
ActiveCN107619873AQuick digRapid phenotypeMicrobiological testing/measurementDNA/RNA fragmentationAmylaseCandidate Gene Association Study
The invention discloses a method for developing waxy1-gene internal molecule markers on the basis of association analysis and KASP. One indel and two snap markers located in waxy1 gene and remarkablyassociated with content of amylase are found out mainly through genome-wide association study and candidate gene association analysis techniques; the detected indel is marked as a major mutant type ofglutinous maize in China; glutinous maize materials can be identified through agarose gel by the markers developed on the basis of the indel, and convenience and rapidness is achieved as compared with that of original polyacrylamide gel identification; meanwhile, paired linkage disequilibrium test of 59 parts of waxy1-gene sequence regions of maize hybrid line materials is performed, close linkage of the indel marker and the two snp markers is tested, primers of the two snps are developed on the basis of the KASP technology, the glutinous maize materials can be identified rapidly, efficientlyand accurately, and molecular breeding of glutinous maize is facilitated.
Owner:SHANGHAI JIAO TONG UNIV
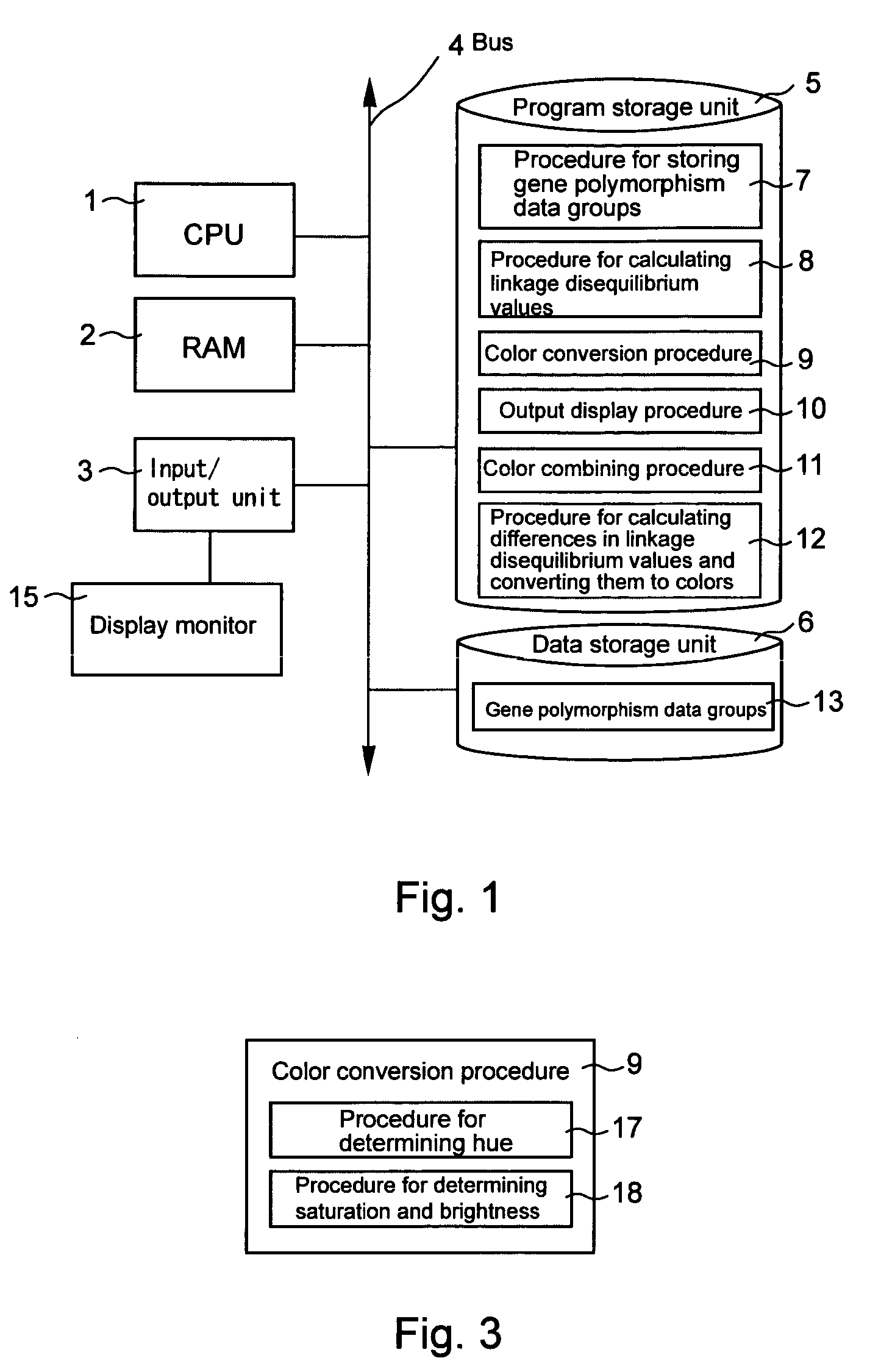
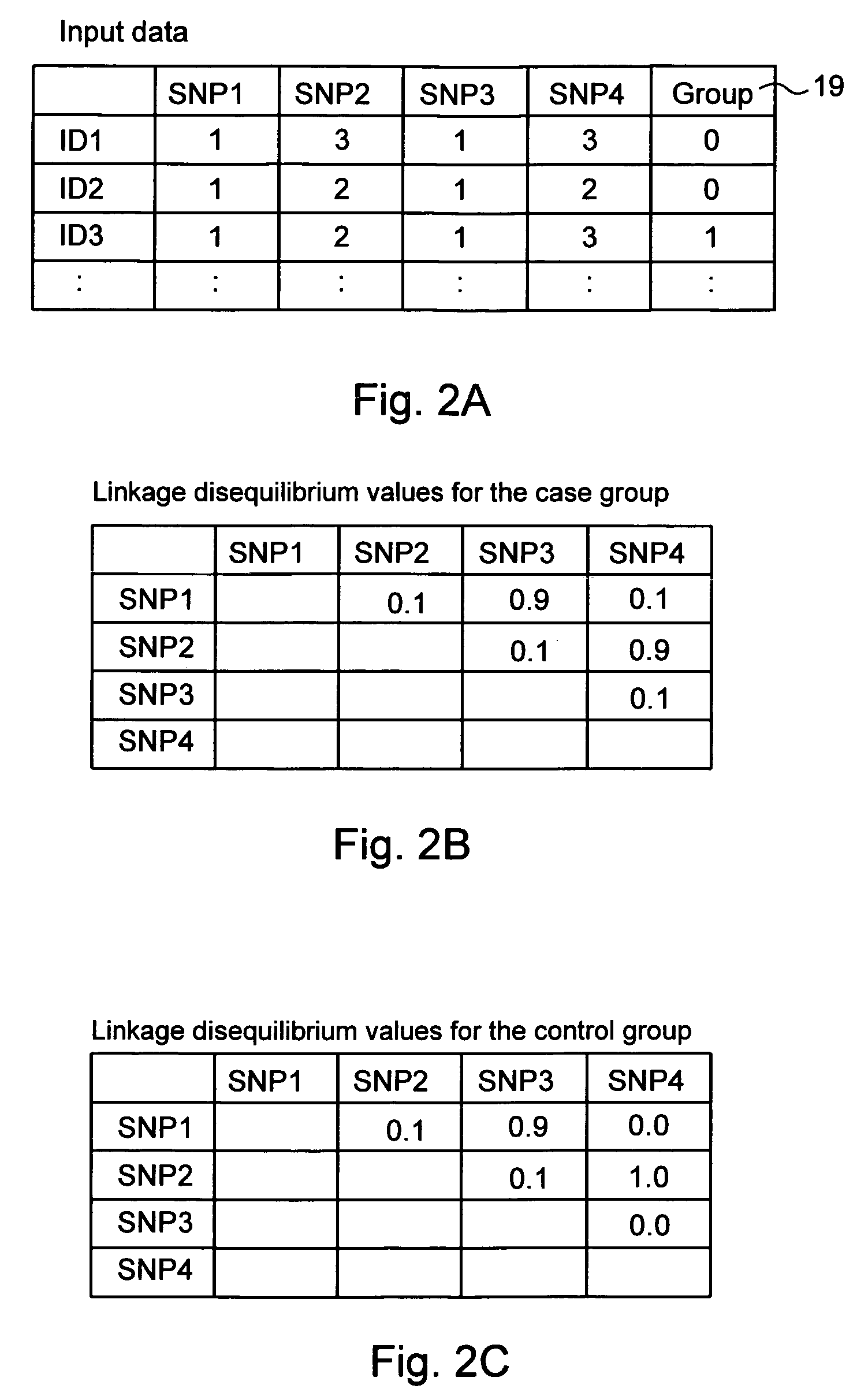
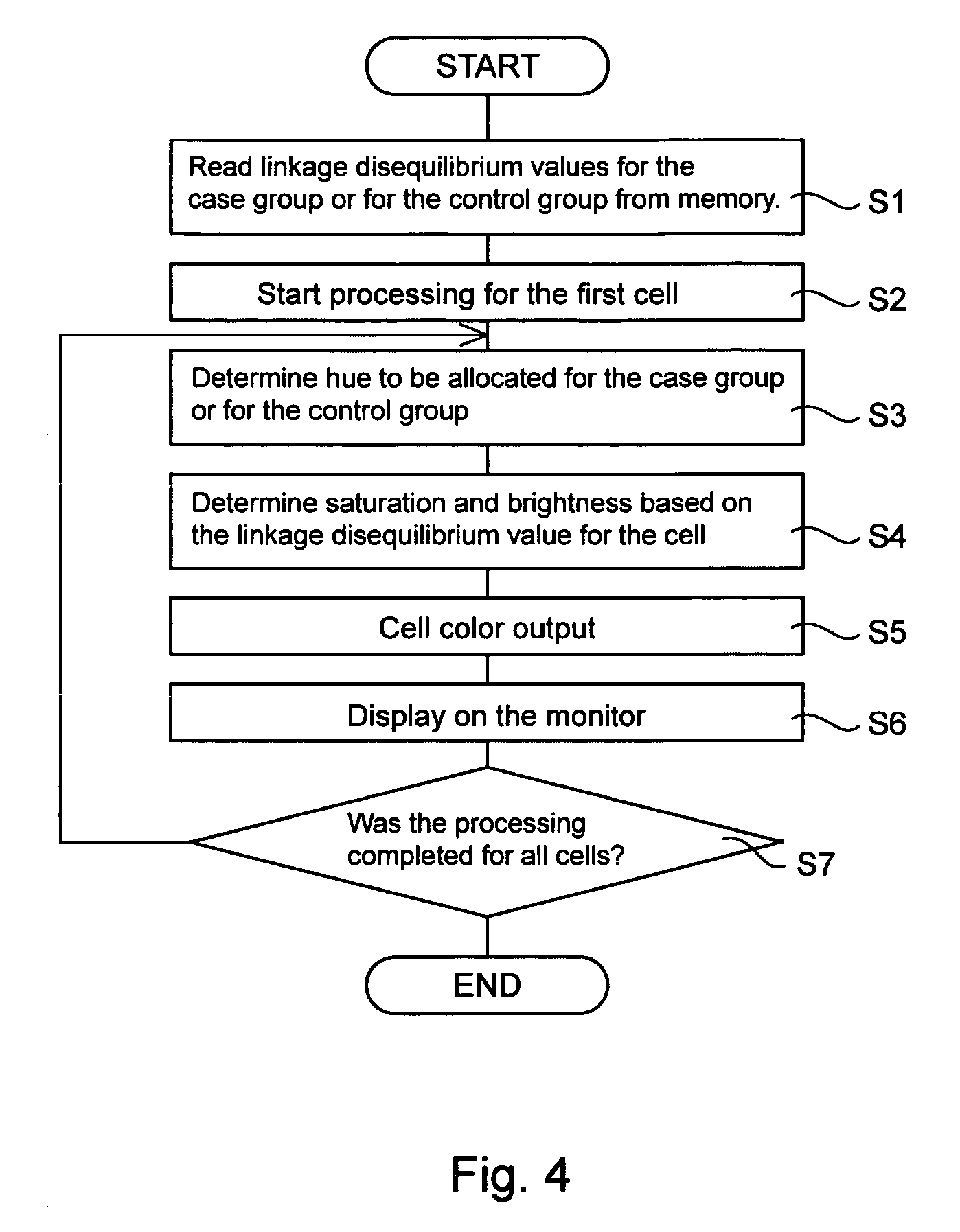
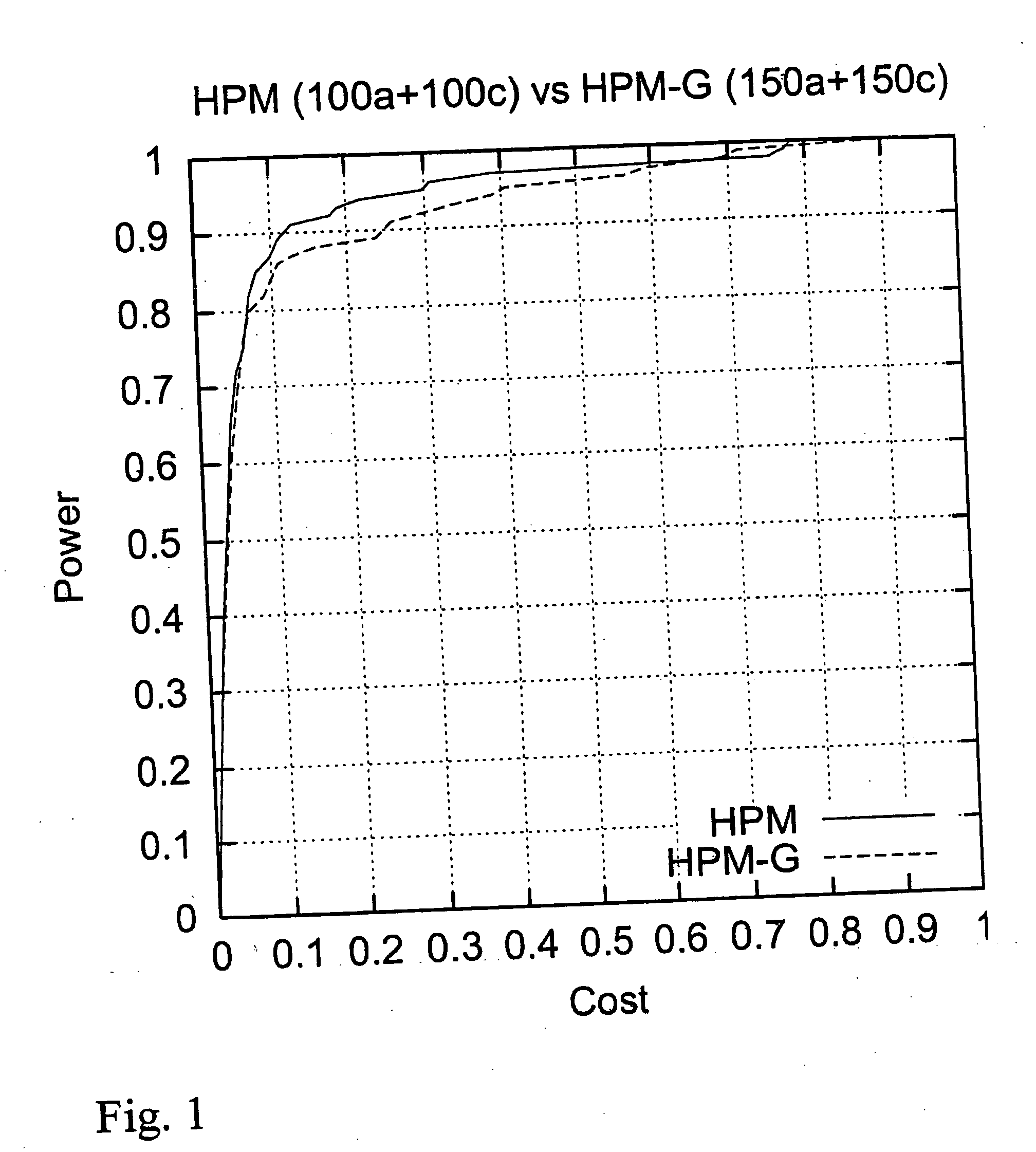
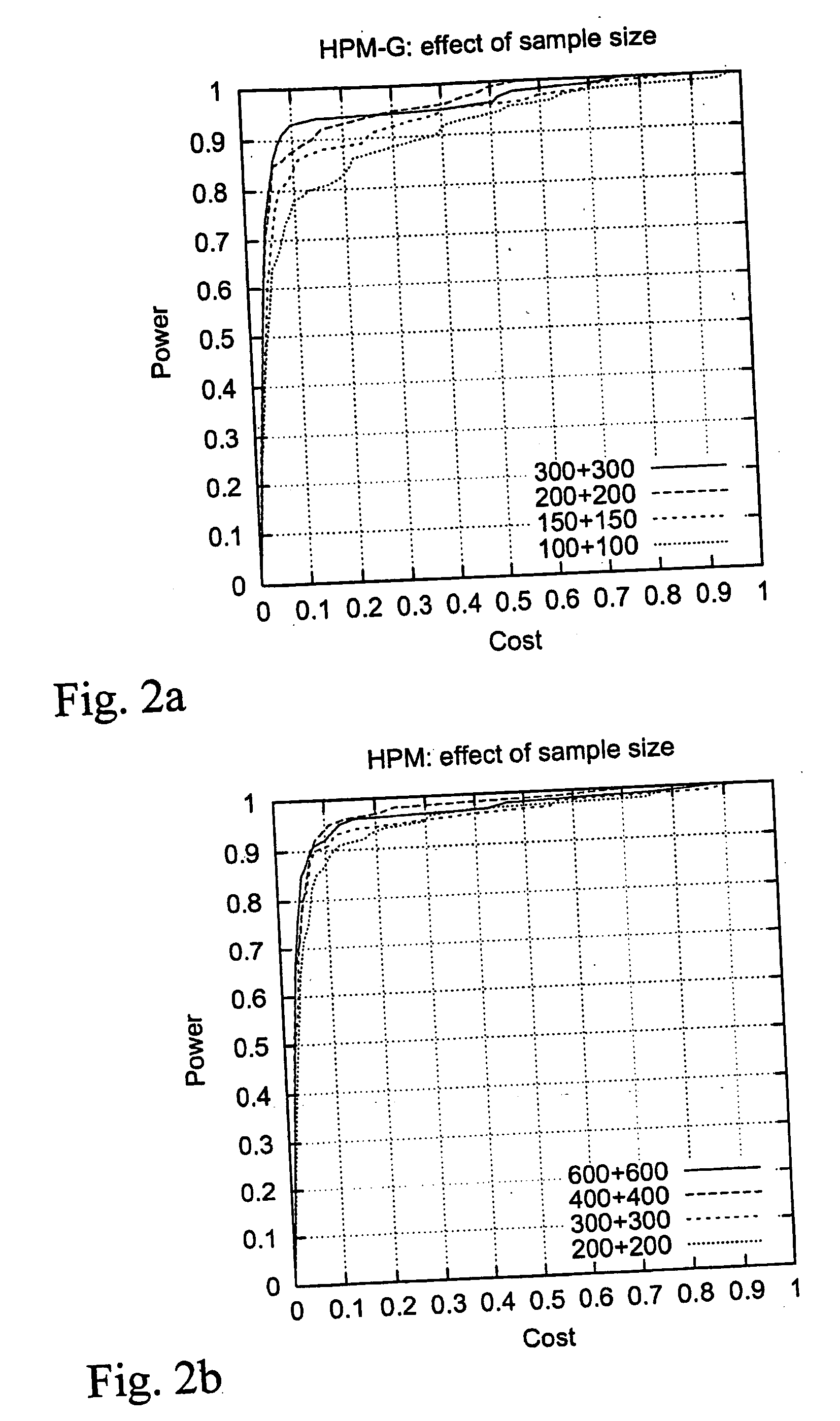
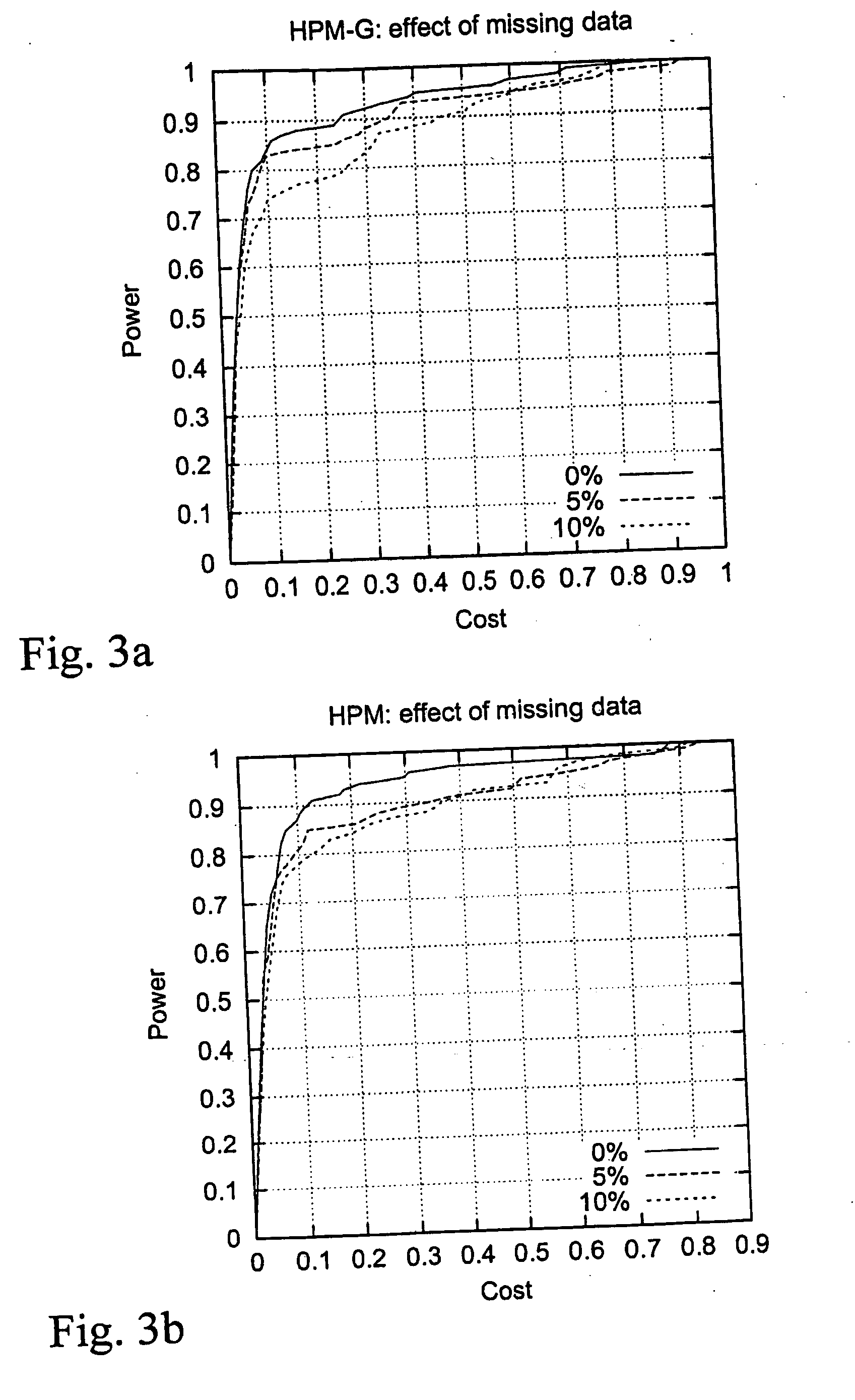
Computer software program for graphically displaying genetic linkage disequilibrium, and the method thereof
InactiveUS20040260479A1Reduce in quantityReduce calculationData visualisationProteomicsGraphicsDensity based
This invention relates to a computer software program product, including computer readable memory and a computer software program stored on the memory, for calculating linkage disequilibrium values for individual pairs of gene loci for two or more gene polymorphism data groups and displaying results comparatively on a display monitor. The program includes: a color output command for converting the linkage disequilibrium values for individual pairs of gene loci for a first gene polymorphism data group and those for a second gene polymorphism data group into a first color set and a second color set, each color set comprising colors with differently allocated saturation, brightness and density based on the linkage disequilibrium values, and for outputting the two color sets; and a comparative display command for displaying comparative results for the first and second color sets on a display monitor in such a way that comparison of the disequilibrium values between the first and second gene polymorphism data groups can be made.
Owner:DYNACOM
Method for gene mapping from genotype and phenotype data
InactiveUS20050250098A1Model-free and computationally effectiveOvercomes drawbackMicrobiological testing/measurementBiostatisticsGene mappingGenetic linkage disequilibrium
A method for gene mapping from genotype and phenotype data utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. All marker patterns P that satisfy a certain pattern evaluation function e(P) are searched from the data, each marker mi of the data is scored by a marker score and the location of the gene is predicted as a function of the scores s(mi) of all the markers mi in the data.
Owner:LICENTIA OY
Method for increasing paddy stigma exsertion rate
InactiveCN107200775AQuick selectionMaintain stable productionMicrobiological testing/measurementPlant peptidesOryzaMolecular breeding
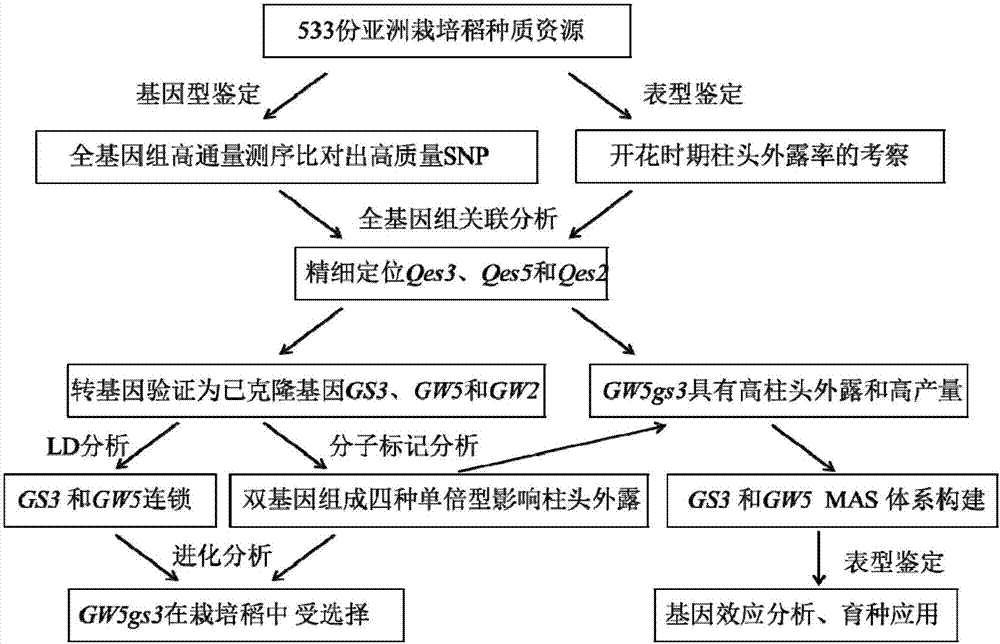
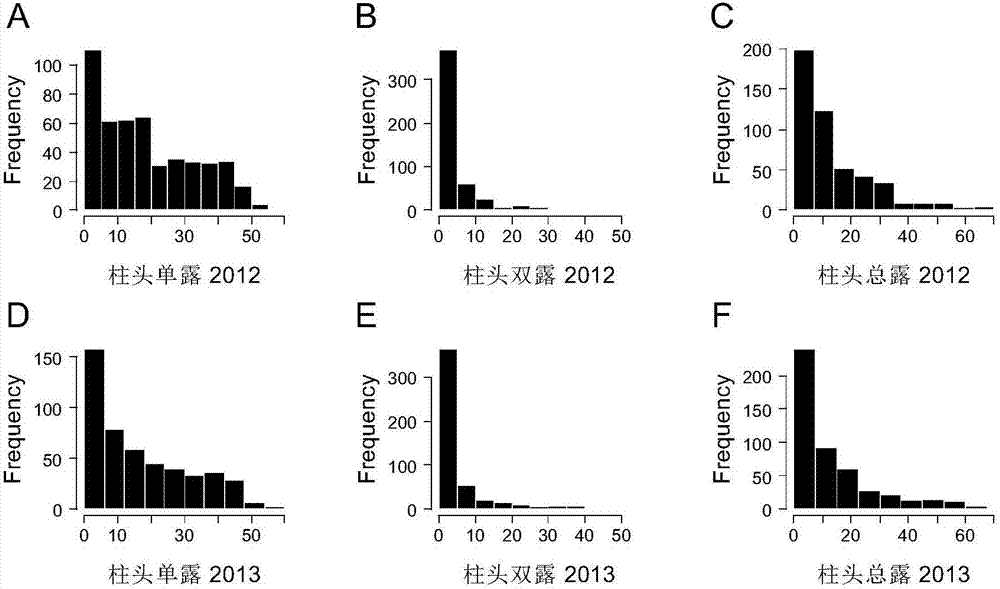
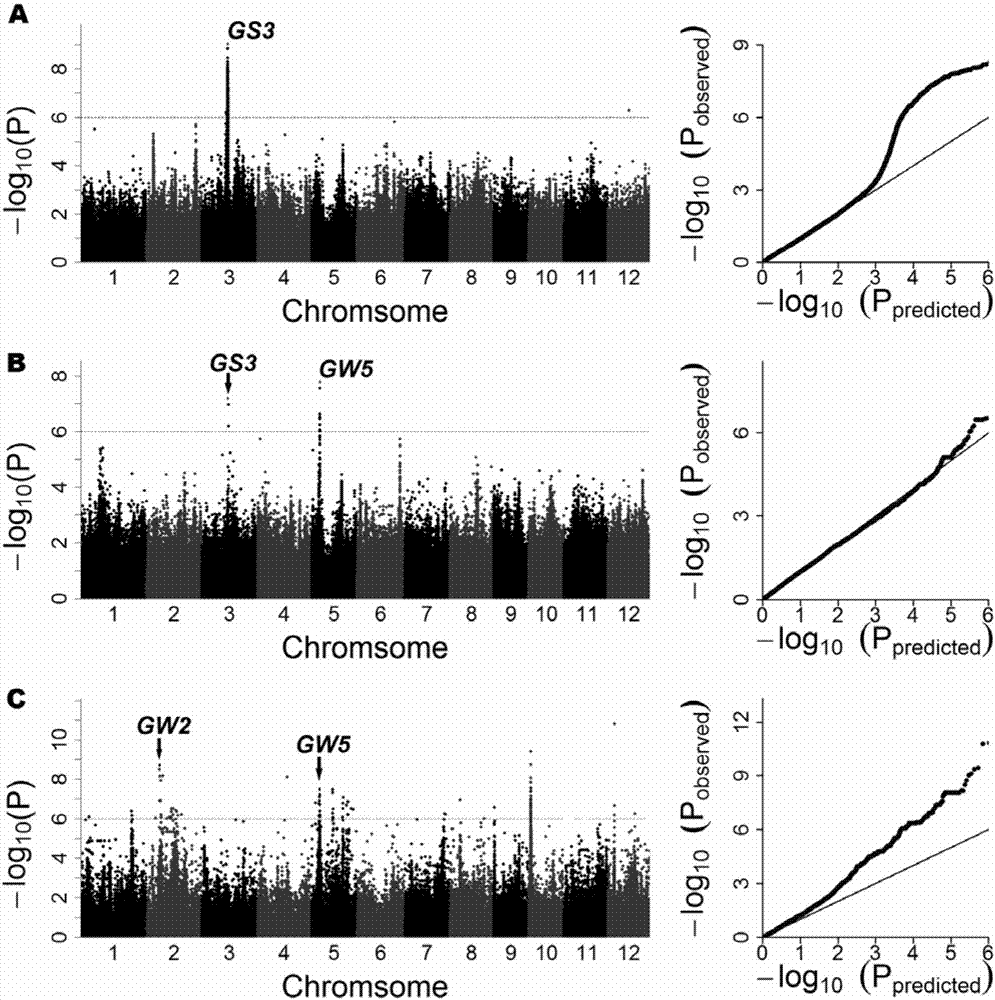
The invention belongs to the technical field of rice molecular breeding. In particular, it relates to a method for increasing the rice stigma exposure rate. The present invention utilizes the GS3 gene dCAPS marker SF28 and the GW5 gene STS marker Indel1 or Indel2 or N1212 to conduct association analysis on 533 cultivated rice genomes, locate 3 QTLs affecting stigma exposure, and confirm that GS3 and GW5 genes are the ones that affect rice The main genes of stigma exposure, GS3 and GW5 genes are in linkage disequilibrium, which affect the rate of stigma exposure in rice by regulating seed size and stigma length.
Owner:HUAZHONG AGRI UNIV
Method for effectively anchoring candidate gene region of peanut quantitative traits
InactiveCN109694924AAchieve positioningMicrobiological testing/measurementBiotechnologyGenetic linkage disequilibrium
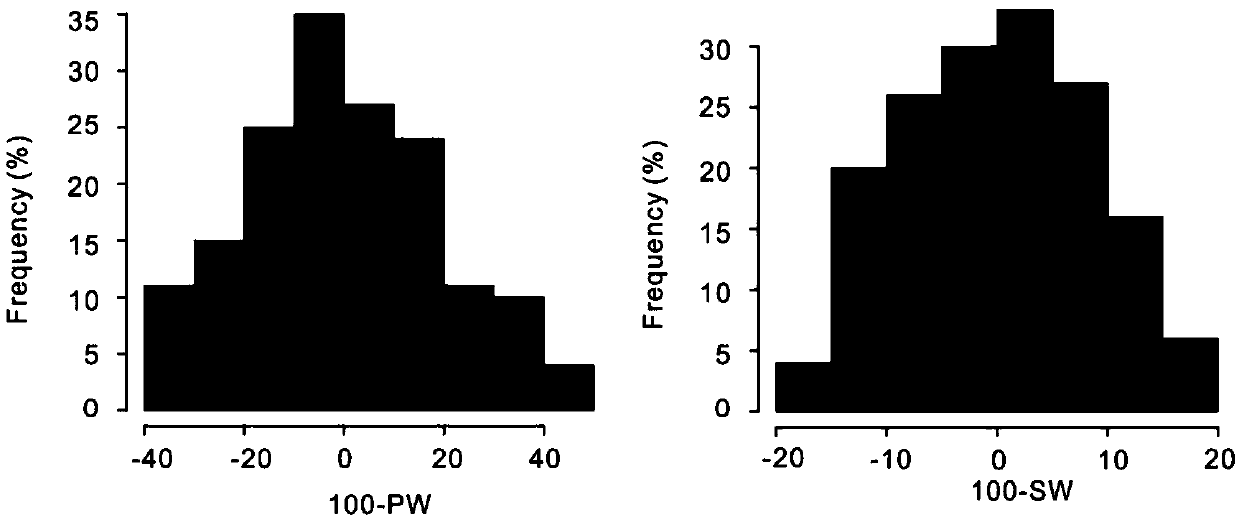
The invention relates to the field of biotechnology, and in particular to a method for effectively anchoring a candidate gene region of a quantitative trait. The method is co-localized with a genome-wide association study and a quantitative trait localization method to achieve efficient anchoring of the quantitative trait candidate gene regions. The method achieves effective anchoring of the candidate gene region of the peanut quantitative trait by co-localization with the genome-wide association study and the quantitative trait localization. The method is exemplified by quantitative granule weight, firstly, through a simplified genome sequencing technology, the genome-wide SNP marker development of 165 cultivar peanut core collections in China is carried out; based on a principle of linkage disequilibrium, combined with peanut 100-fruit weight and 100-kernel weight breeding value, the related gene regions related to peanut weight traits are associated; in the previous reports, the QTLmapping results of the grain weight traits are converted to the corresponding genomic physical locations, the co-localization region of the two methods can be obtained, and the method can effectivelyand accurately realize the localization of the genome-wide particle weight-related trait genes.
Owner:SHANDONG PEANUT RES INST
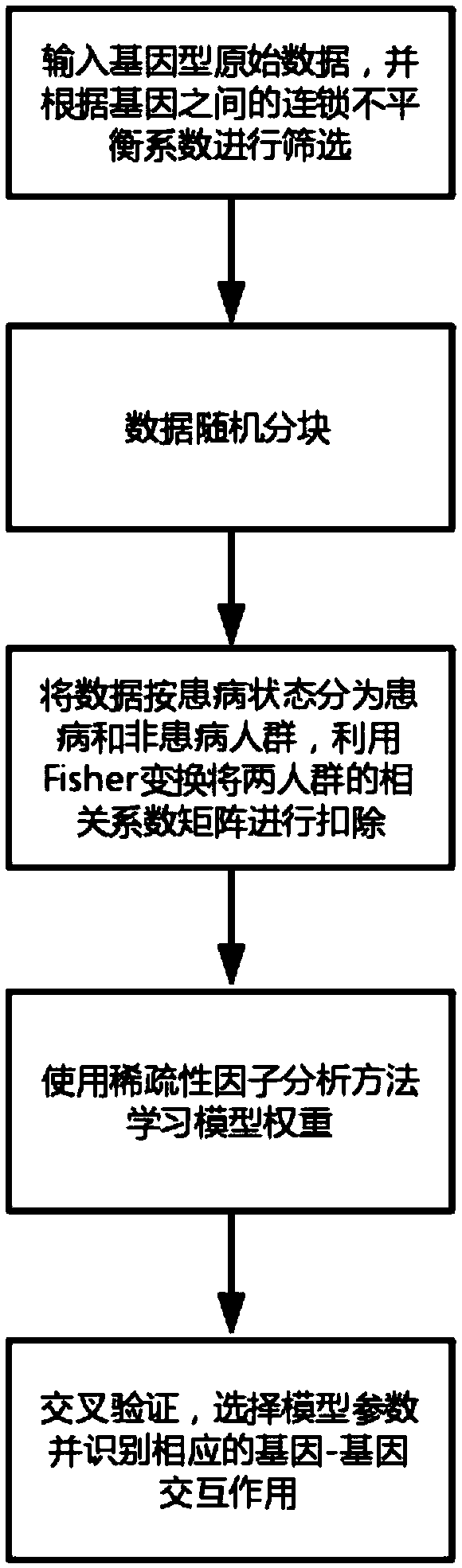
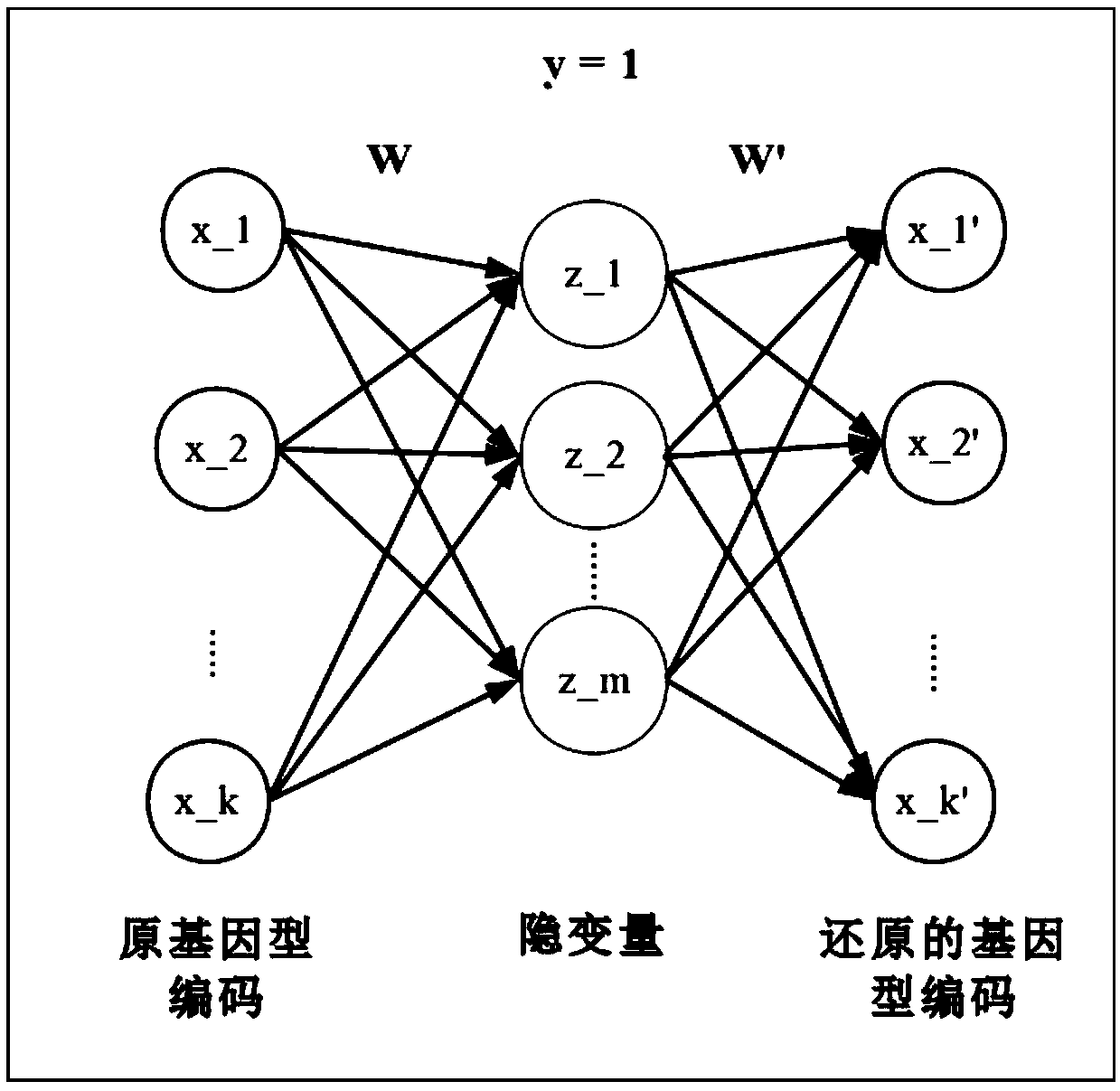
Gene-gene interaction recognition method based on sparsity factor analysis
The invention discloses a gene-gene interaction recognition method based on the sparsity factor analysis (Sparse Factor Analysis for Epistasis, EPISFA). The method comprises the following steps of 1)entering genotype raw data and screening according to a linkage disequilibrium coefficient between genes; 2) randomly dividing the data into blocks; 3) dividing the data into diseased and non-diseasedgroups according to a disease state, calculating the correlation coefficient matrix of two groups, and using Fisher transform to deduct gene site correlation of the correlation coefficient matrix ofthe two groups; 4) learning model weights using a sparsity factor analysis method; 5) conducting cross-validation, selecting model parameters, and identifying corresponding gene-gene interaction. Experiments show that the statistical efficiency and computational efficiency of the method are both high and the method has good application prospects.
Owner:PEKING UNIV
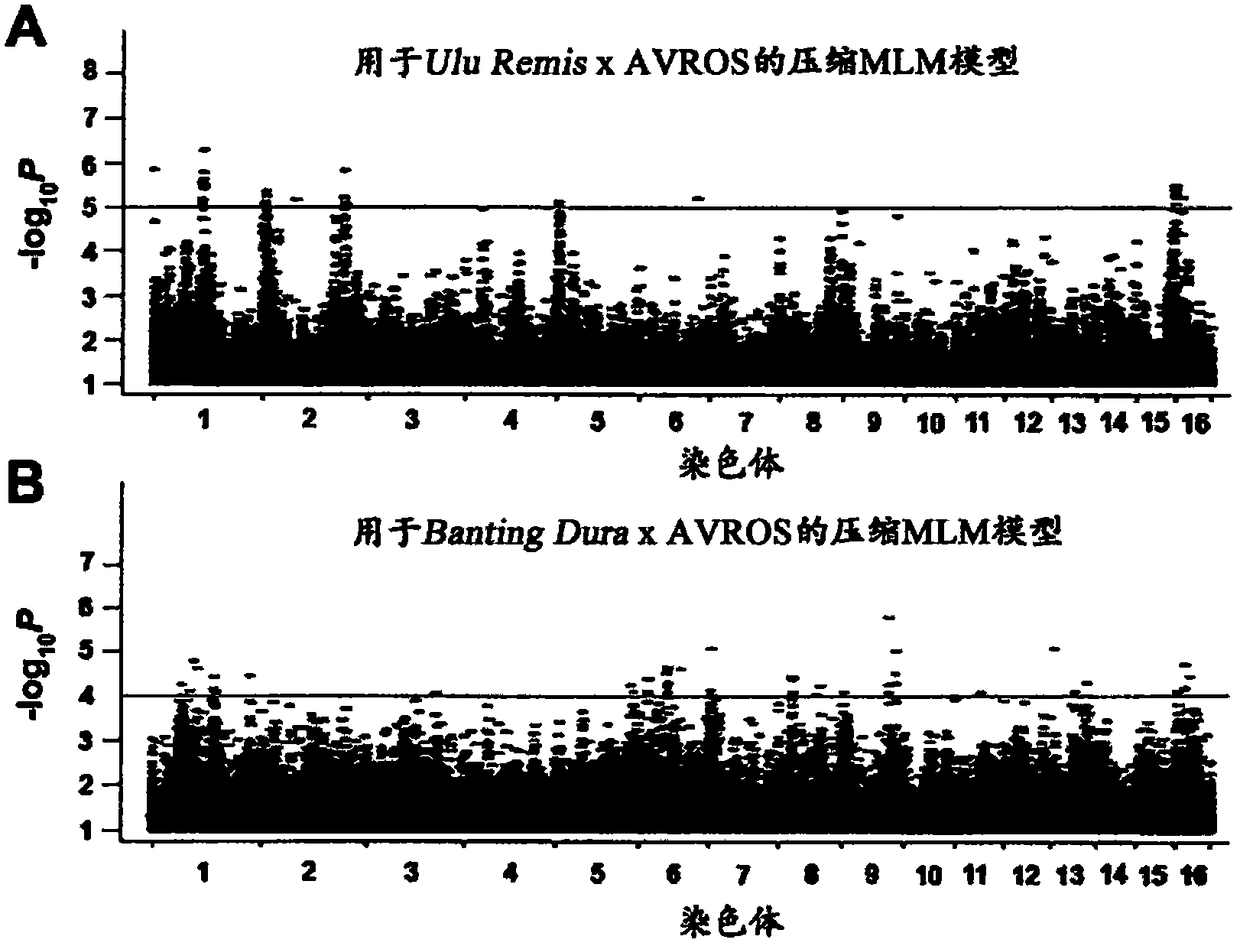
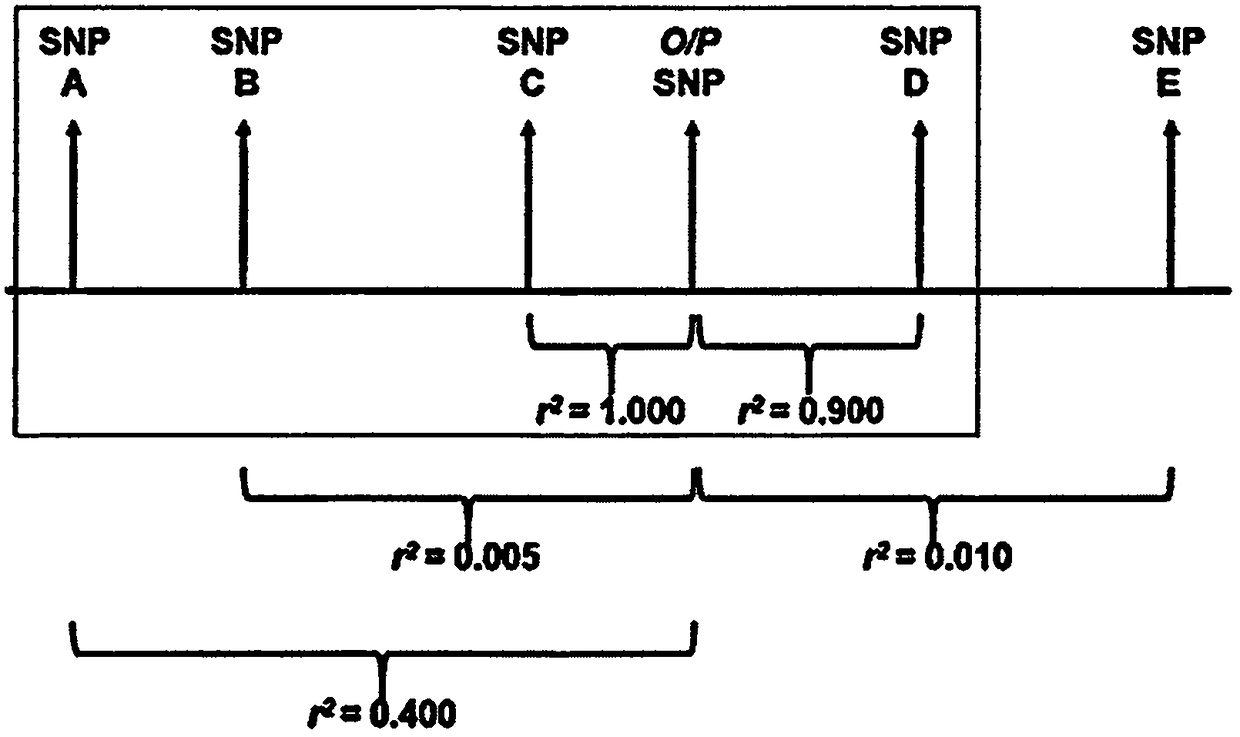
Methods and SNP detection kits for predicting palm oil yield of a test oil palm plant
Methods for predicting palm oil yield of a test oil palm plant are disclosed. The methods comprise determining, from a sample of a test oil palm plant of a population, at least a first SNP genotype, corresponding to a first SNP marker, located in a first QTL for a high-oil- production trait and associated, after stratification and kinship correction, with the high-oil- production trait with a genome-wide -log10(p-value) of at least 4.0 in the population or having a linkage disequilibrium r2 value of at least 0.2 with respect to a first other SNP marker linked thereto and associated, after stratification and kinship correction, with the high-oil-production trait with a genome-wide -log10(p-value) of at least 4.0 in the population. The methods also comprise comparing the first SNP genotype to a corresponding first reference SNP genotype and predicting palm oil yield of the test plant based on extent of matching of the SNP genotypes.
Owner:SIME DARBY PLANTATION INTPROP SDN BHD
Method for gene mapping from chromosome and phenotype data
InactiveUS20050064408A1Microbiological testing/measurementProteomicsGene mappingGenetic linkage disequilibrium
The present invention relates to a method for gene mapping from chromosome and phenotype data, which utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. The method according to the invention is based on discovering and assessing tree-like patterns in genetic marker data. It extracts, essentially in the form of substrings and prefix trees, information about the historical recombinations in the population. This infor-mation is used to locate fragments potentially inherited from a common diseased founder, and to map the disease gene into the most likely such fragment. The method measures for each chromosomal location the disequilibrium of the prefix tree of marker strings starting from the location, to assess the distribution of disease-associated chromosomes.
Owner:LICENTIA OY
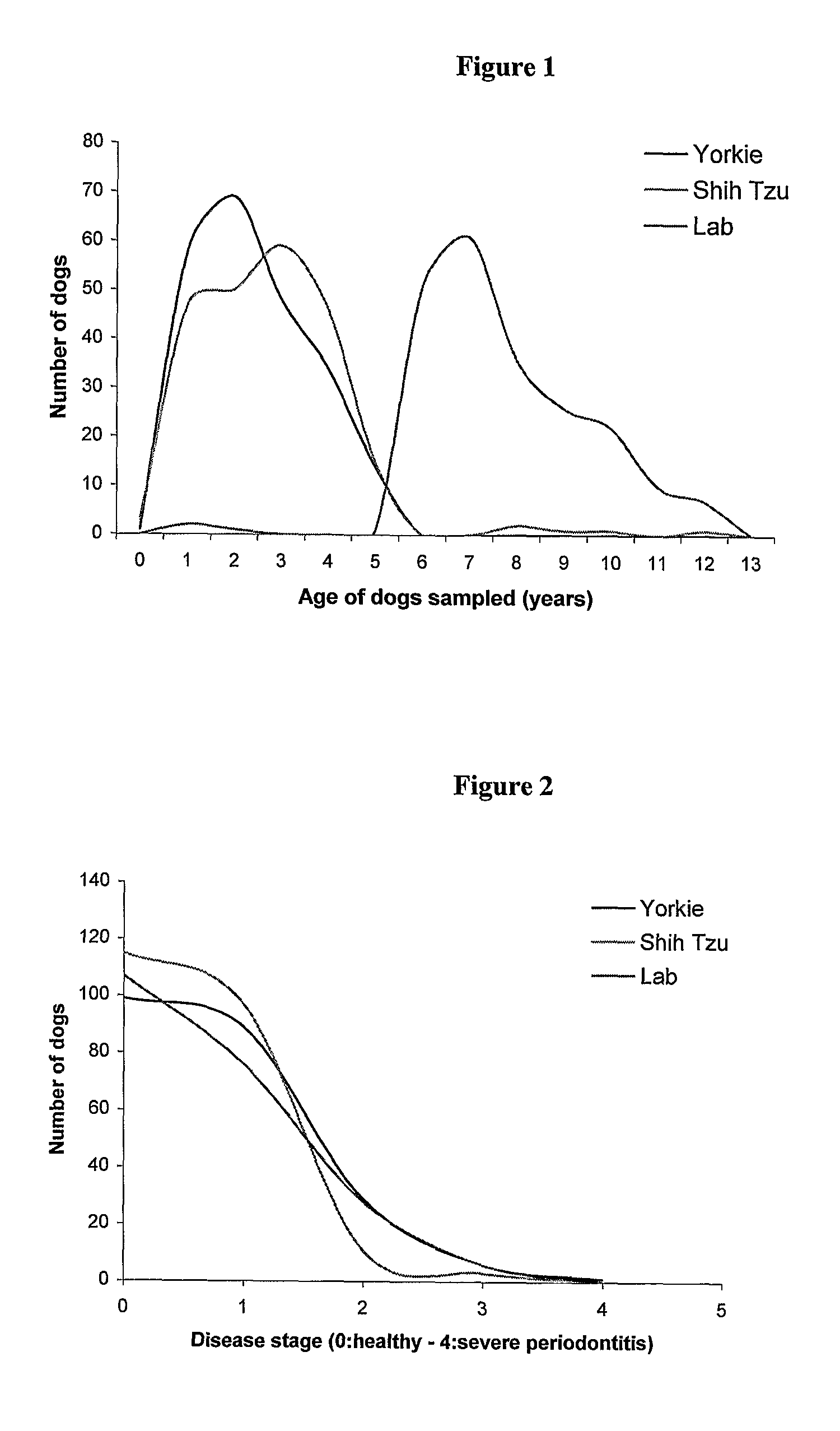
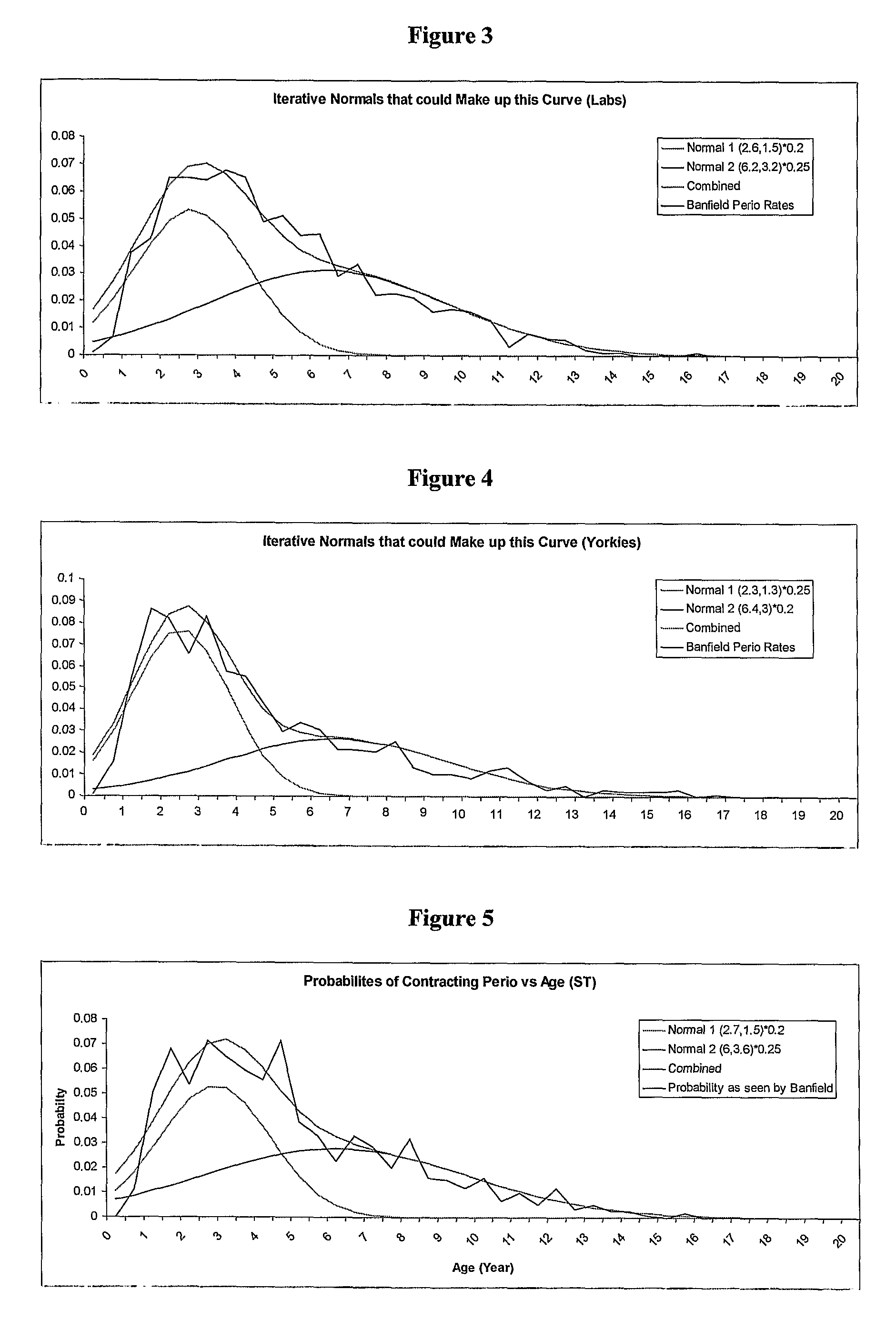
Dog Periodontitis
InactiveUS20080226766A1Delay and prevent onsetSusceptibility to periodontitisBacterial antigen ingredientsMicrobiological testing/measurementNucleotideLinkage disequilibrium
A method of determining susceptibility to periodontitis in a Shih Tzu dog, Yorkshire Terrier dog or a dog of a breed that is genetically related to the Shih Tzu or Yorkshire Terrier breed, comprises: a) typing the nucleotide present in the genome of the dog at or at a position equivalent to each of the following: position 201 of SEQ ID NO: 2 (SNP_02), or a position that is in linkage disequilibrium with this position,—position 201 of SEQ ID NO: 4 (SNP_04), or a position that is in linkage disequilibrium with this position, and position 201 of SEQ ID NO: 9 (SNPJ39), or a position that is in linkage disequilibrium with this position, and b) thereby determining whether the dog is susceptible to periodontitis.
Owner:MARS INC
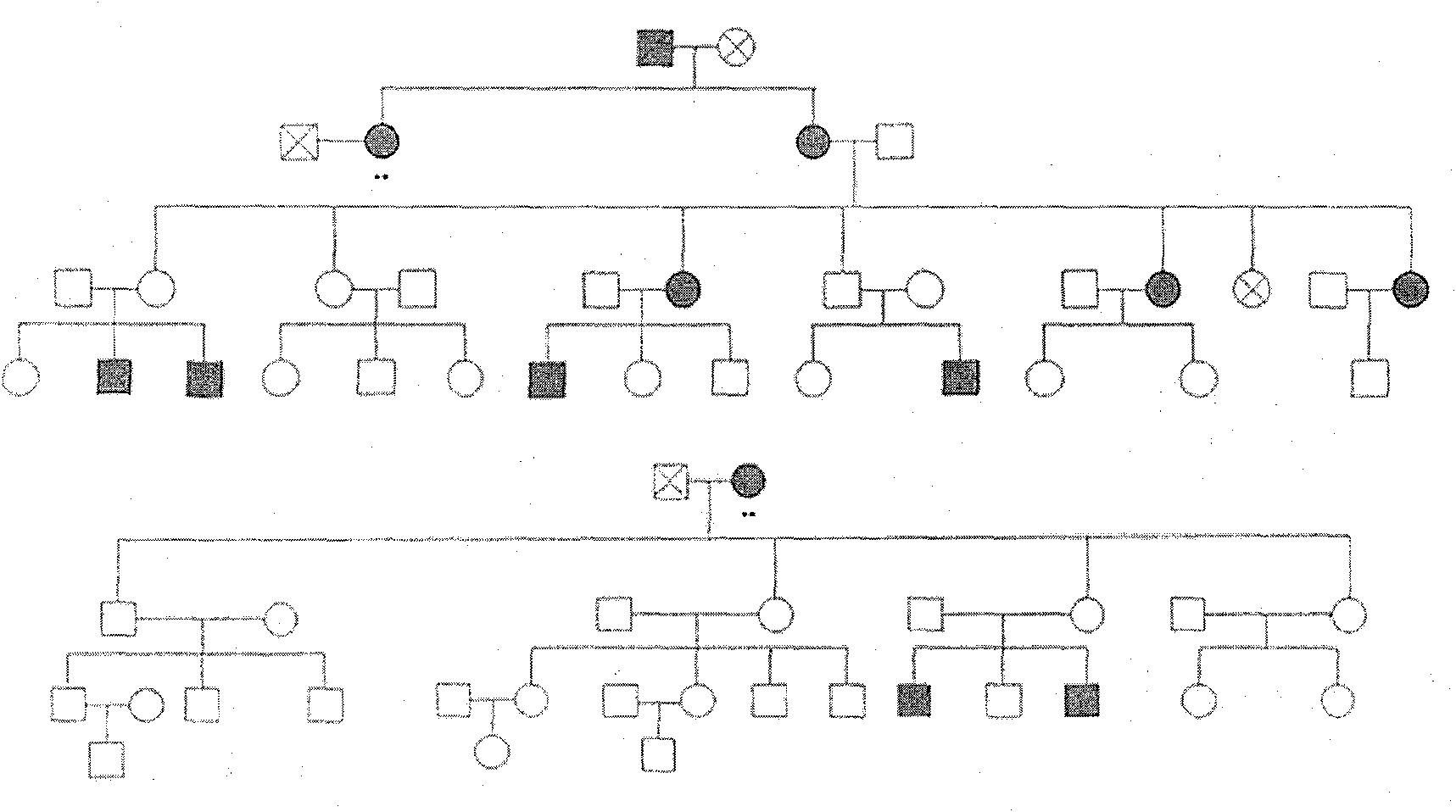
Method for screening Wilson disease gene mutation in family by using linkage disequilibrium
ActiveCN103820555ALow costMicrobiological testing/measurementThree levelLinkage Disequilibrium Mapping
The invention discloses a method for screening Wilson disease gene mutation in a family by using linkage disequilibrium, which is based on a linkage disequilibrium principle, by a mononucleotide polymorphism genetic typing method, six mononucleotide polymorphism sites are used for determining genotype state of hepatolenticular degeneration patients and its brothers and sisters (or relatives in three levels), and then a haplotype of other members in family is compared with a haplotype of a propositus for determining the sufferers or heterozygosis carriers.
Owner:李卫东
SSR markers for plants and uses thereof
Simple sequence repeat (SSR) markers identified in Jatropha curcas and useful for the molecular genotyping of plants. are described. These markers may be used for identifying allele polymorphisms, identifying identical or related plants, differentiating plants and studying genetic diversity in a population. The markers may also be used in genetic and phenotype studies using statistical methods, for example, linkage analysis, association mapping, linkage disequilibrium and the like. The information may be used for breeding and / or selection of plants.
Owner:ACGT INTELLECTUAL
Methods for predicting palm oil yield of a test oil palm plant
Owner:SIME DARBY PLANTATION INTPROP SDN BHD
SNP molecular markers, primer pairs and kit of FABP4 gene related to beef quality in Yanbian yellow cattle and application of SNP molecular marker, primer pair and kit
ActiveCN110438243AMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
The invention provides SNP molecular markers, primer pairs and a kit of FABP4 gene related to beef quality in Yanbian yellow cattle, and application of the SNP molecular markers, the primer pairs andthe kit, and belongs to the technical field of beef quality screening in yellow cattle. The SNP molecular markers of the FABP4 gene contain a nucleotide sequence of a polymorphism at the 3496 bp as anA / C site, a polymorphism at the 3533 bp as an A / T site, a polymorphism at the 3711 bp as a G / C site, a polymorphism at the 3745 bp as a T / C site and a polymorphism at the 3767 bp as a T / C site of theFABP4 gene. The above five polymorphic sites are located in the third exon, and are in a strong linkage disequilibrium state with similar genetic effects. When the SNP molecular markers are applied to screen breeding fat cattle, fat content and marble pattern are selected as the detection indicators for screening breeding fat cattle while the genotype is heterozygous.
Owner:YANBIAN UNIV
Method for predicting the athletic performance potential of a subject
ActiveUS20140189894A1Reduce operating costsEnhance the imageMicrobiological testing/measurementAvicultureExercise performanceGenetics
A method for predicting the athletic performance potential of a subject comprising the step of assaying a biological sample from a subject for a genetic variant in linkage disequilibrium with MSTN-66493737 (T / C) SNP. The invention also provides an assay for determining the athletic performance potential of a subject.
Owner:UNIV COLLEGE DUBLIN NAT UNIV OF IRELAND DUBLIN
Prognosis of response to treatment with Anti-tnf-alpha in patients with rheumatoid arthritis
The invention relates to the use of SNP rs3794271, and / or an SNP that is in total linkage disequilibrium with same, as a marker in predicting the response to treatment with anti-TNF in a patient with RA. The invention also relates to methods for predicting the response to treatment with anti-TNF, as well as for deciding on or recommending a treatment for a patient with RA, based on determining the genotype for rs3794271 and / or an SNP that is in total linkage disequilibrium with same.
Owner:FUNDACIO HOSPITAL UNIVERSITARI VALL DHEBRON INST DE RECERCA
Method for assisting prediction of risk of occurrence of side effect of irinotecan
ActiveCN107208163AMicrobiological testing/measurementRecombinant DNA-technologySide effectGenetic linkage disequilibrium
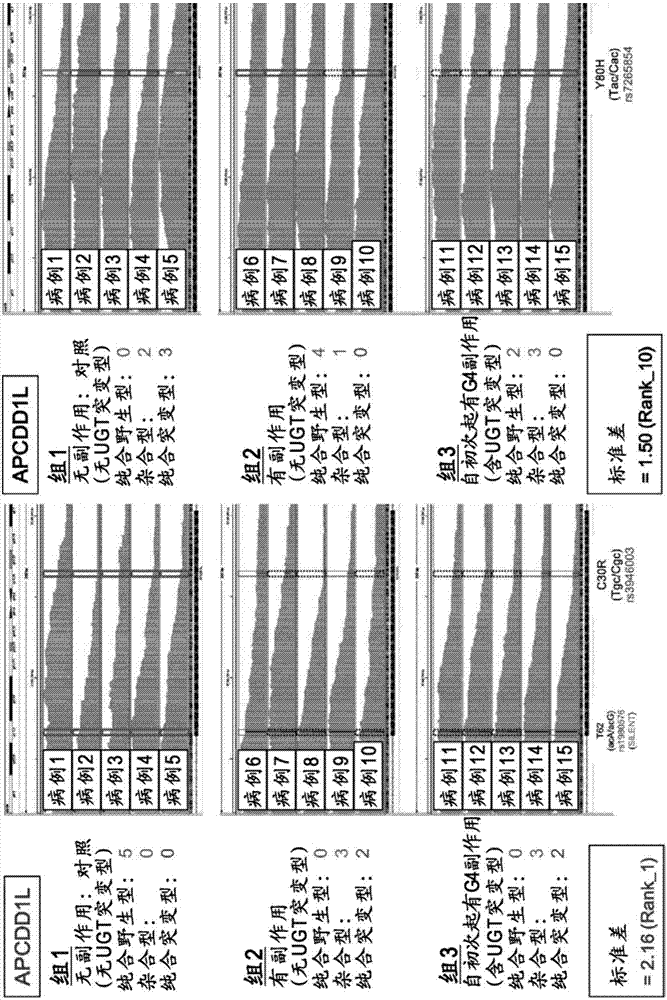
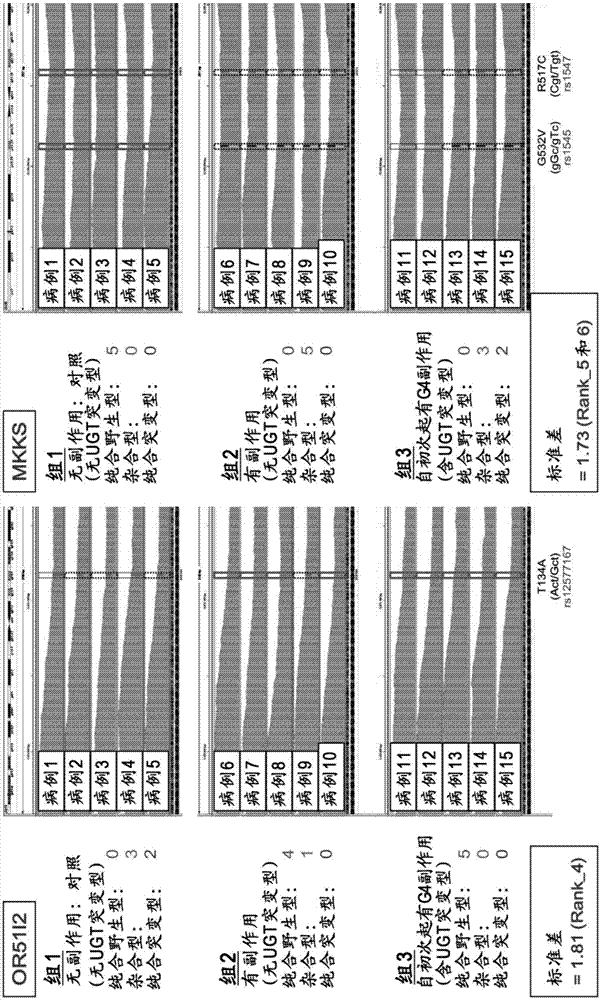
The present invention addresses the problem of providing a simple and efficient means for predicting the risk of occurrence of a side effect of irinotecan by analyzing a single nucleotide polymorphism in a region encoding a specific gene. The prediction of the risk of occurrence of a side effect of irinotecan is assisted by analyzing: a single nucleotide polymorphism in a region encoding the APCDD1L gene, the R3HCC1 gene, the OR51I2 gene, the MKKS gene, the EDEM3 gene, or the ACOX1 gene which are present on genomic DNA in a biological sample collected from a subject; or a single nucleotide polymorphism which is in linkage disequilibrium with or genetically linked to the single nucleotide polymorphism, thereby determining whether the single nucleotide polymorphism is homozygous or heterozygous for a variant-type, or homozygous for a wild-type and assisting prediction of the risk of occurrence of a side effect of irinotecan.
Owner:YAMAGUCHI UNIV +1
Genetic test and pet diet
InactiveUS20100278938A1Preventing liver copper accumulationReducing hepatic copper concentrationBiocideInorganic active ingredientsDiseaseLabrador Retriever
The present invention provides a method of determining the susceptibility of a dog to liver copper accumulation comprising detecting the presence or absence of (a) a polymorphism in the GOLGA5, ATP7a or UBL5 gene of the dog that is indicative of susceptibility to copper accumulation and / or (b) a polymorphism in linkage disequilibrium with a said polymorphism (a), and thereby determining the susceptibility of the dog to liver copper accumulation. The invention also provides a foodstuff comprising copper at a concentration of less than 21 mg / kg dry matter for use in preventing a disease attributable to liver copper accumulation in a dog having genetic inheritance of the Labrador Retriever breed.
Owner:MARS INC
Chromosomal Blocks as Markers for Traits
InactiveUS20090246774A1Sugar derivativesMicrobiological testing/measurementBiotechnologyGenetic linkage disequilibrium
The present invention provided a method for predicting a phenotype in a bovine animal, the method comprising analysing a nucleic acid sample from said animal for the presence of at least one genetic marker known to reside in an Linkage Disequilibrium (LD) block in any one of bovine chromosomes BTA-I to BTA-29, wherein said LD block is associated with said phenotype. The phenotype can be Australia profit ranking (APR), Australian selection index (ASR), protein yield (PROT), protein percent (PROT %), milk volume (MILK), fat yield (FAT), fat percent (FAT %), breeding value overall type (Overall Type), somatic cell count (SCC), and / or breeding value cow fertility (Cow Fertility). Also provided is a linkage disequilibrium unit (LDU) map of any one or more of bovine chromosomes BTA-I to BTA-29, the map comprising a plurality of chromosomal regions, and the regions defined by their co-inheritance across generations substantially as entire linkage disequilibrium (LD) blocks.
Owner:INNOVATIVE DAIRY PRODS TRUSTEE FOR THE PARTICIPANTS OF THE COOP RES CENT FOR INNOVATIVE DAIRY PRODS
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com