Method for effectively anchoring candidate gene region of peanut quantitative traits
A technology of candidate genes and quantitative traits, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of lack of in-depth analysis of peanut characteristics and their relationship, and achieve the effect of precise and fine interval positioning
Inactive Publication Date: 2019-04-30
SHANDONG PEANUT RES INST
View PDF3 Cites 6 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment Construction
[0038] In order to facilitate those skilled in the art to understand the present invention, the specific implementation of the present invention will be described below in conjunction with the examples.
[0039] 1. Sample collection and DNA extraction
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
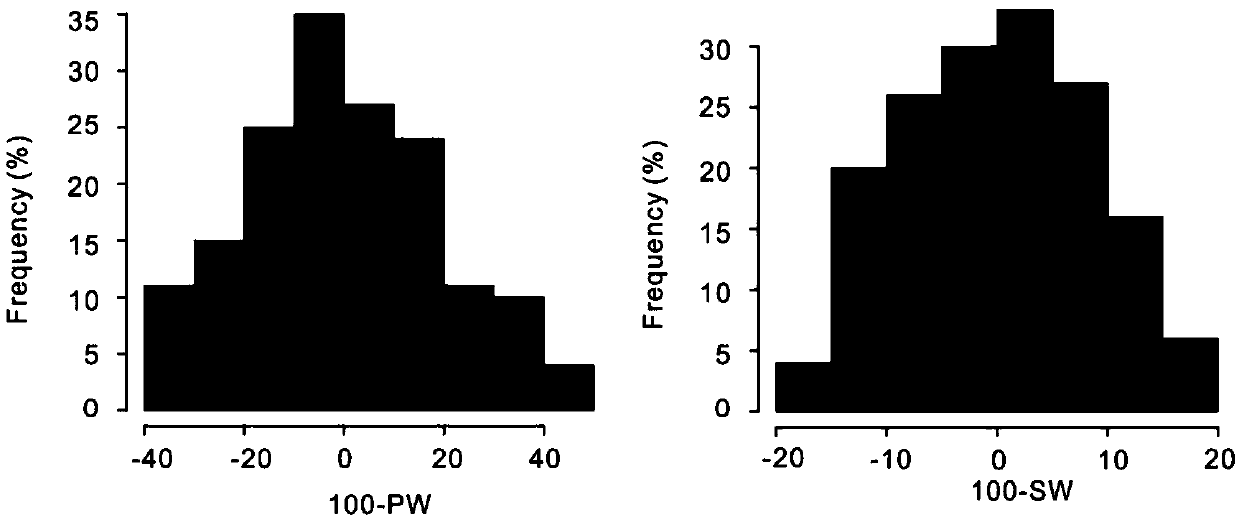
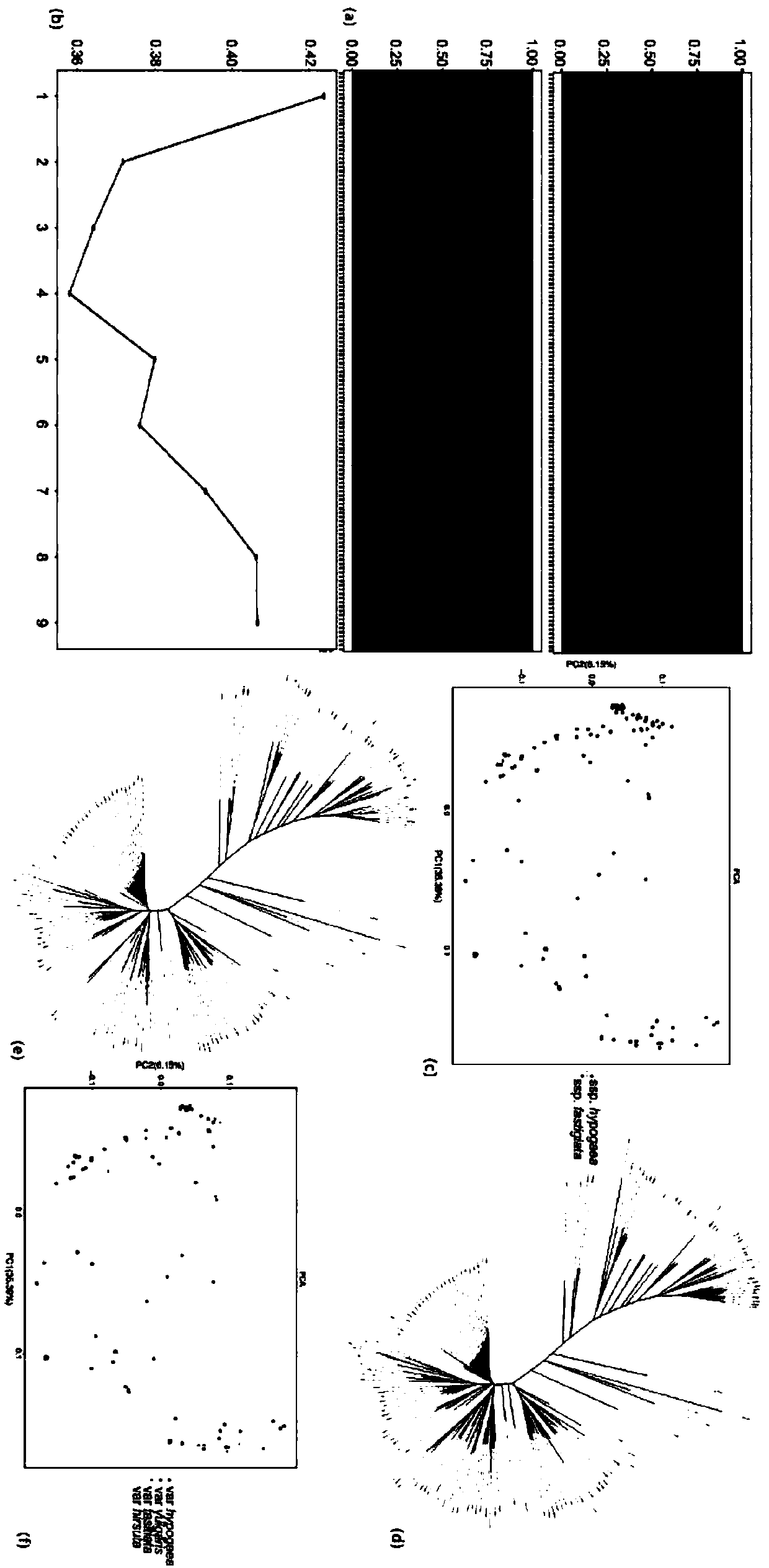
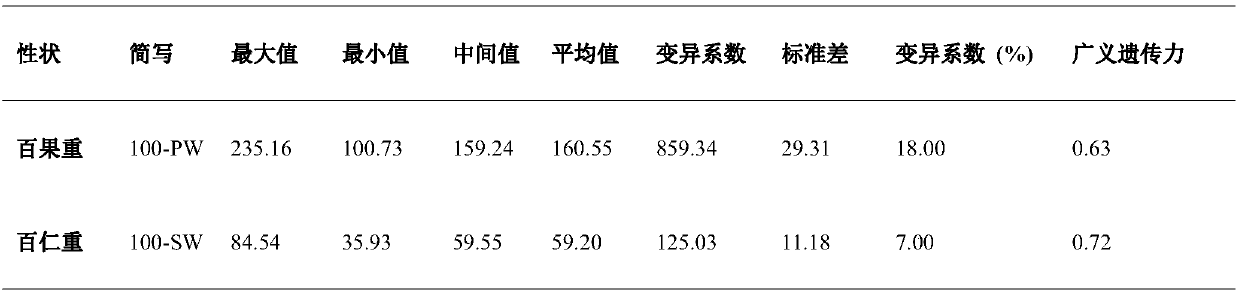
The invention relates to the field of biotechnology, and in particular to a method for effectively anchoring a candidate gene region of a quantitative trait. The method is co-localized with a genome-wide association study and a quantitative trait localization method to achieve efficient anchoring of the quantitative trait candidate gene regions. The method achieves effective anchoring of the candidate gene region of the peanut quantitative trait by co-localization with the genome-wide association study and the quantitative trait localization. The method is exemplified by quantitative granule weight, firstly, through a simplified genome sequencing technology, the genome-wide SNP marker development of 165 cultivar peanut core collections in China is carried out; based on a principle of linkage disequilibrium, combined with peanut 100-fruit weight and 100-kernel weight breeding value, the related gene regions related to peanut weight traits are associated; in the previous reports, the QTLmapping results of the grain weight traits are converted to the corresponding genomic physical locations, the co-localization region of the two methods can be obtained, and the method can effectivelyand accurately realize the localization of the genome-wide particle weight-related trait genes.
Description
technical field [0001] The invention relates to the field of biotechnology, in particular to a method for effectively anchoring candidate gene regions of peanut quantitative traits. Background technique [0002] The peanut cultivar (Arachis hypogaea L.) is an important oil crop worldwide. China is the largest producer of cultivated peanuts, accounting for more than 40% of total global production. However, it still cannot satisfy the national demand for peanuts. Therefore, cultivating high-quality and high-yielding peanut varieties is still a long-term and arduous task. With the development of biotechnology, genetic breeding has avoided the time-consuming and labor-intensive shortcomings of traditional breeding. The premise of genetic breeding is to accurately anchor the relevant candidate gene regions of important quantitative traits such as yield and quality. The aggregation breeding based on favorable alleles related to each trait has become the most concerned hotspot ...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/6895C12Q1/6858C12Q1/6869
CPCC12Q1/6858C12Q1/6869C12Q1/6895C12Q2600/13C12Q2600/156C12Q2535/122
Inventor 王娟闫彩霞单世华李春娟赵小波苑翠玲孙全喜
Owner SHANDONG PEANUT RES INST
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com