Major SNP (single nucleotide polymorphism) marker influencing growth traits of pigs and application thereof in genetic improvement of productivity of breeding pigs
A growth trait and marker technology, which is applied in the fields of animal molecular biotechnology and animal genetics and breeding, and can solve the problems of unidentified major genes and molecular markers.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
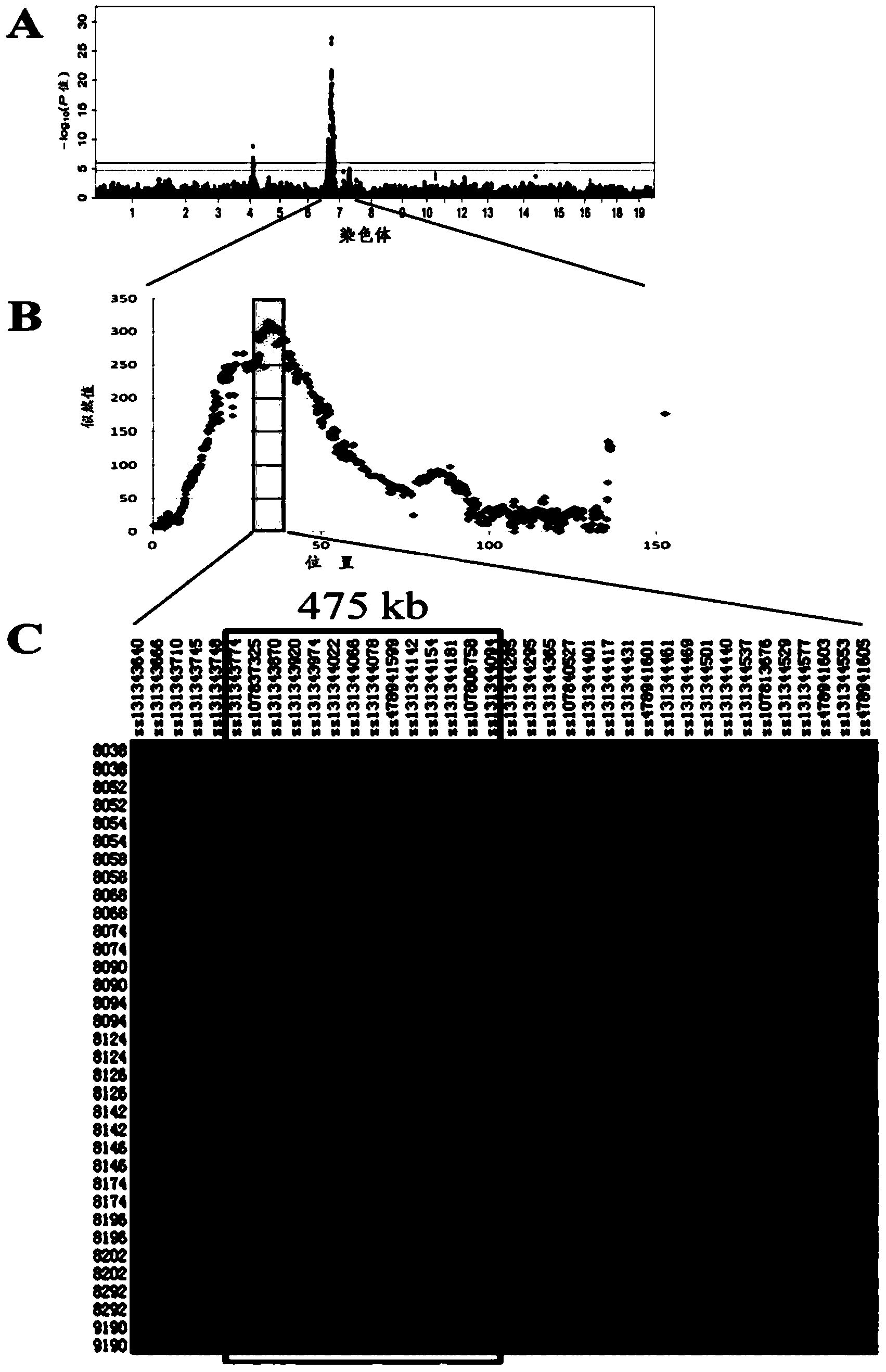
[0019] This example is to specifically explain the inventive process of obtaining the gene marker in the present invention.
[0020] Detection of 60K SNP genotype in the whole genome of pigs: A small piece of ear tissue sample was collected from each individual of the above two experimental groups, genomic DNA was extracted by standard phenol-chloroform method, and the DNA was dissolved in TE buffer. White Duroc × Erhualian pig F 2 The DNA of the resource population was tested for quality and concentration. The A260 / 280 ratio was 1.8-2.0, and the A260 / 230 ratio was 1.7-1.9. Qualified DNA samples were uniformly diluted to 50ng / μL -1 , using the Illumina Infinium SNP typing platform, order the Porcine SNP60DNA Analysis Kit chip, perform chip hybridization and result scanning according to the Illumina Infinium instructions and standard procedures, and read the genotype data through GenomeStudio software. The quality control of the obtained genotype data was carried out with PLI...
Embodiment 2
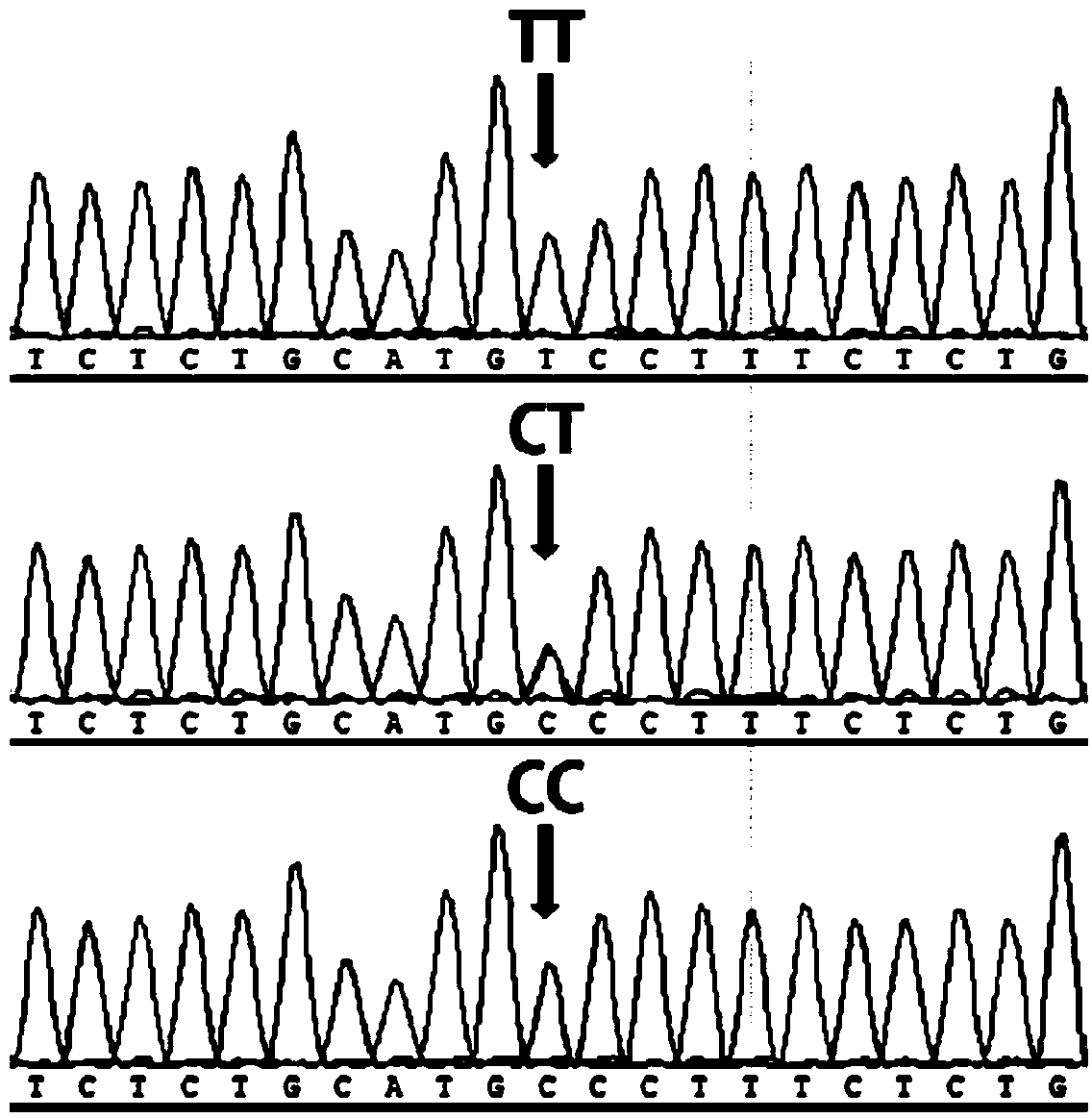
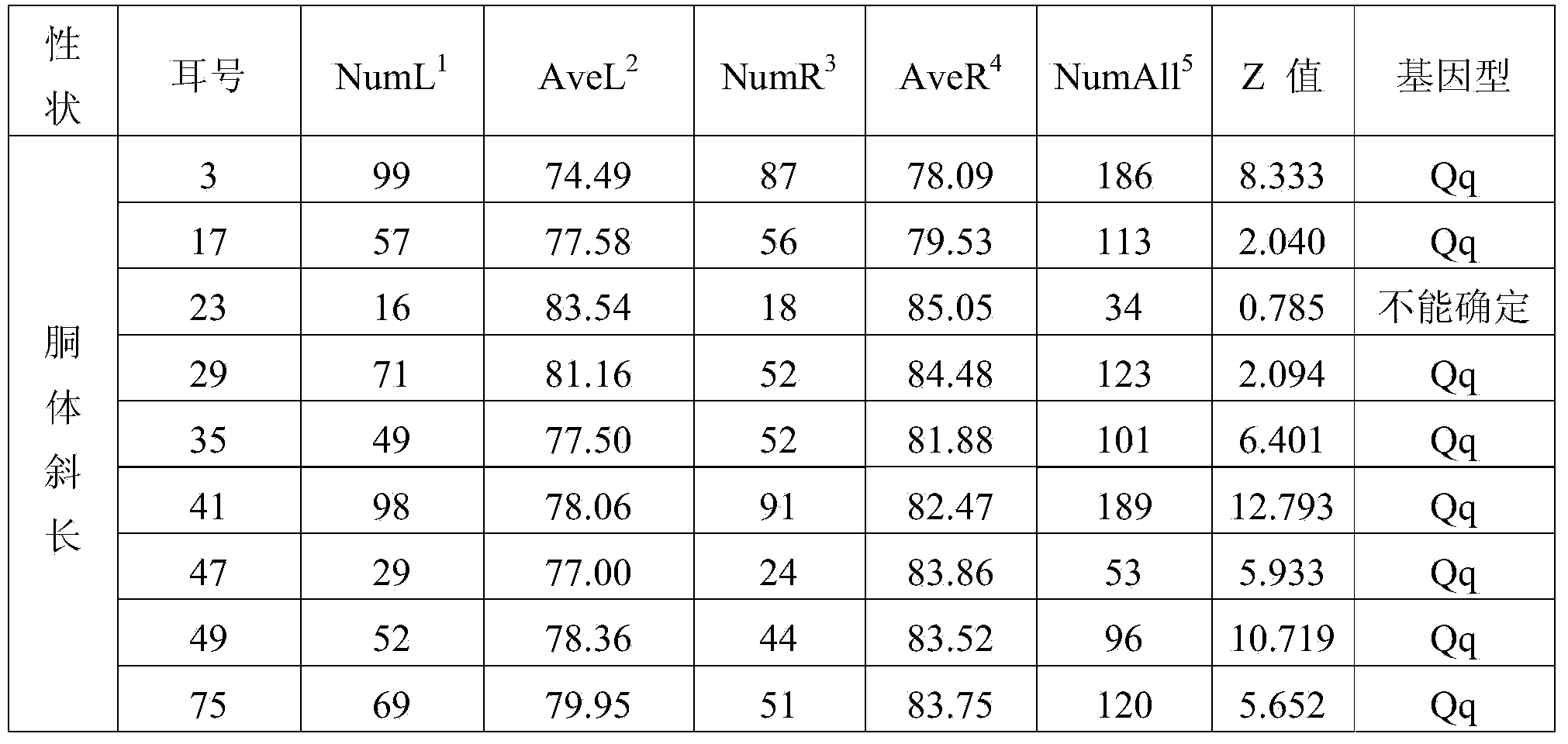
[0042] This example is the application of the SNP site g.34986810C>T obtained in Example 1 in breeding pigs. According to E Q Analysis of the distribution frequency of haplotypes in Chinese local pig breeds. For the core population of Chinese local pigs and the synthetic and matching lines of Chinese local pigs, the above-mentioned method of direct sequencing after PCR amplification was used to detect pig No. 7 The main mutation site g.34986810C>T genotype on the chromosome that affects the growth traits of pigs, select the favorable genotype TT to reserve seeds for individuals, and increase the live body length, live body height, carcass length, and Growth performance such as carcass weight, daily gain and head weight.
[0043] In this embodiment, the Erhualian group is used as experimental animals. Genomic DNA of 334 Erhualian pigs was directly sequenced after PCR amplification with specific primers. The genotype of g.34986810C>T main marker locus was determined according...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com