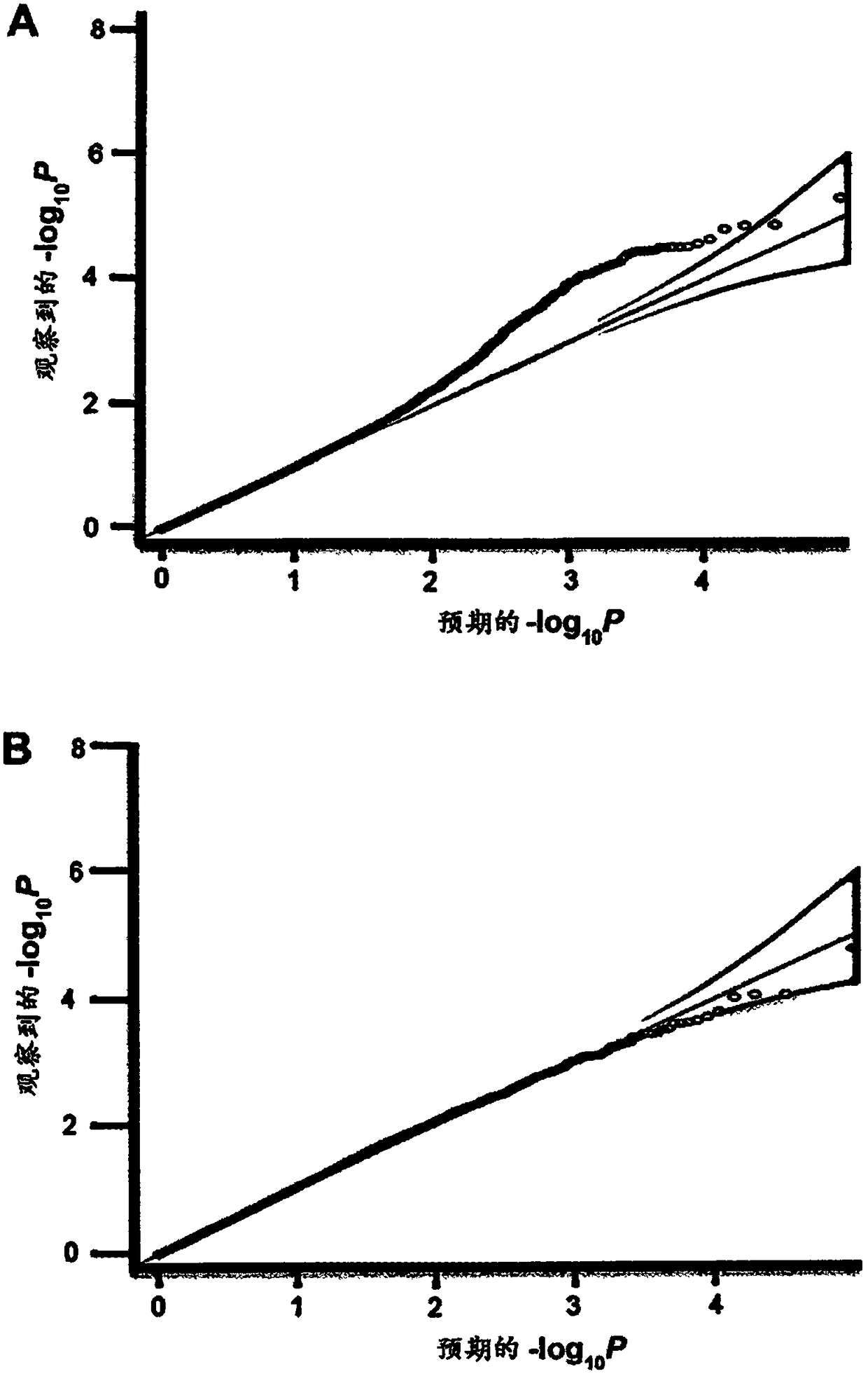
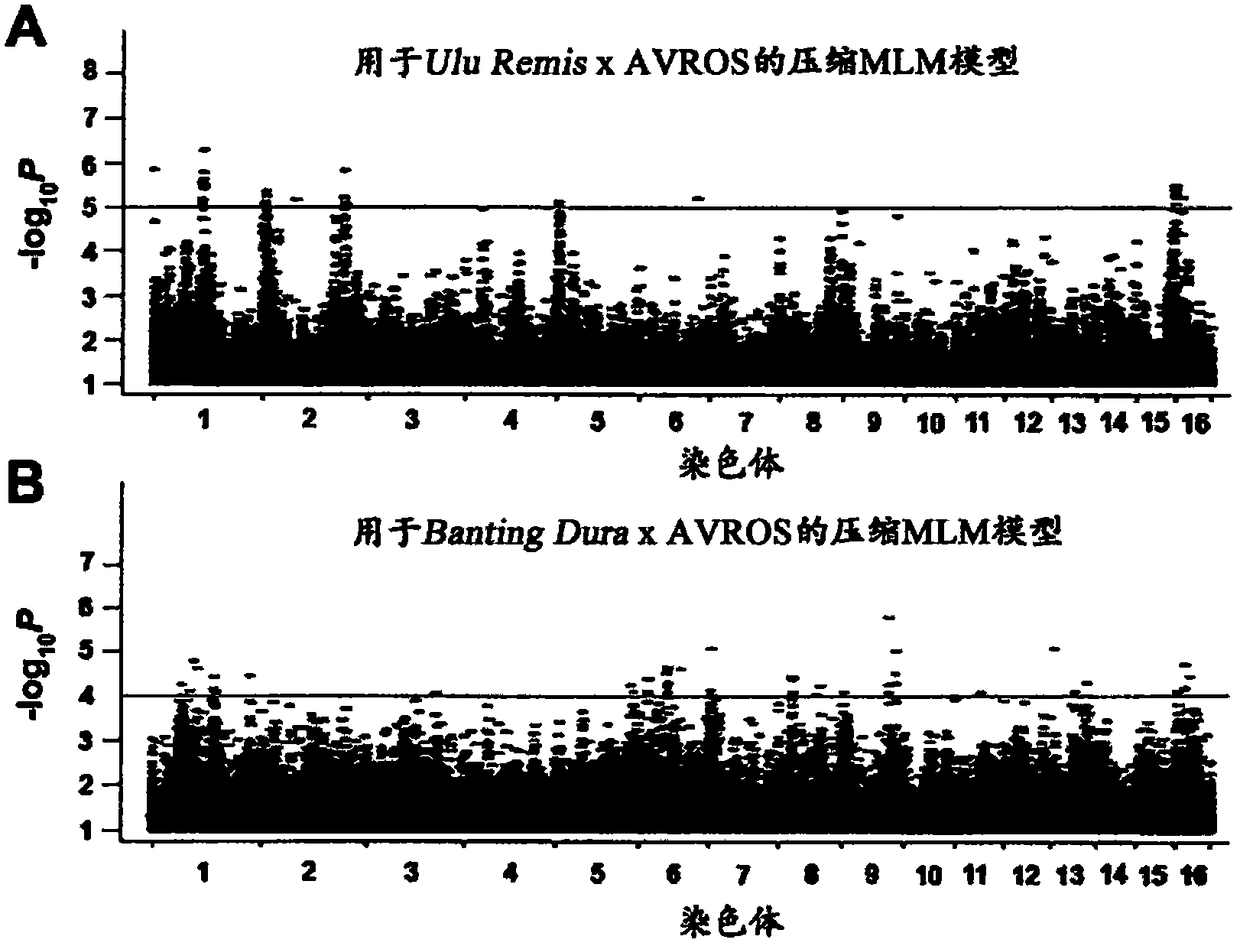
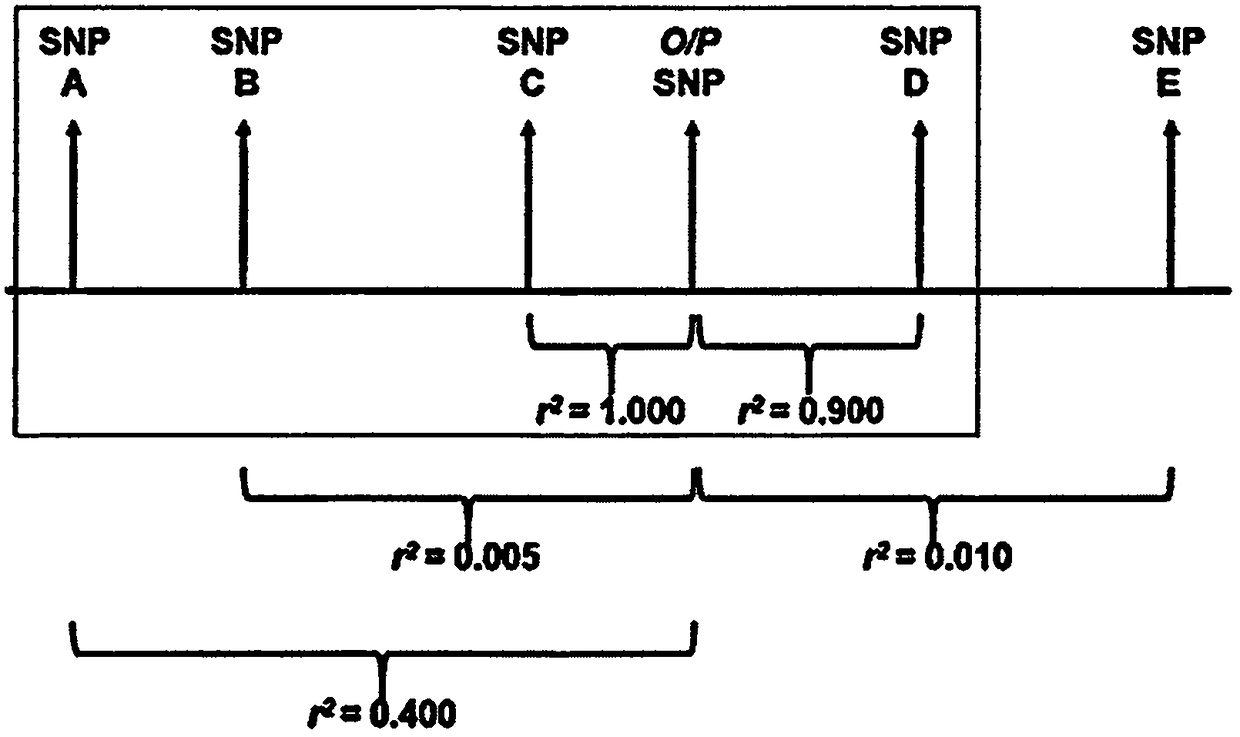
Methods for predicting palm oil yield of a test oil palm plant
A technology for experimental oil and palm oil, which can be used in biochemical equipment and methods, microbial determination/inspection, bioinformatics, etc., and can solve problems such as narrow breeding materials
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0207] Sampling and DNA preparation
[0208] Genome-wide association study (also known as GWAS) mapping populations derived from the Ulu Remis dura x AVROS pisifera population (1,218 palms) and the Banting durax AVROS pisifera population (953 palms) were sampled. Sample selection was based on good representatives of the oil / plant palm plant (also known as O / P) variant and pedigrees recorded by the corresponding breeders. Total genomic DNA was isolated from unexpanded young leaves using the DNAeasy(R) Plant Mini Kit (Qiagen, Limburg, The Netherlands).
[0209] whole genome resequencing
[0210] The samples were pooled based on equal molar concentrations of DNA from each sample to form a sequencing DNA pool. Resequenced libraries were prepared using the HiSeq2000(TM) Sequencing System (Illumina, San Diego, CA) to generate 100 bp paired-end reads to 35x genome coverage, resulting in 1,015,758,056 raw reads. Using BWAMapper (published as Li & Durbin, Bioinformatics 26: 589-595 ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com