Chinese population linkage analysis single nucleotide polymorphism (SNP) marker sets and use method and application thereof
A linkage analysis, a Chinese technology, applied in the field of linkage analysis in genetics and genomics, can solve the problems of difficult product application and inability to meet the genetic analysis of Asian populations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1. The method of using the SNP marker site:
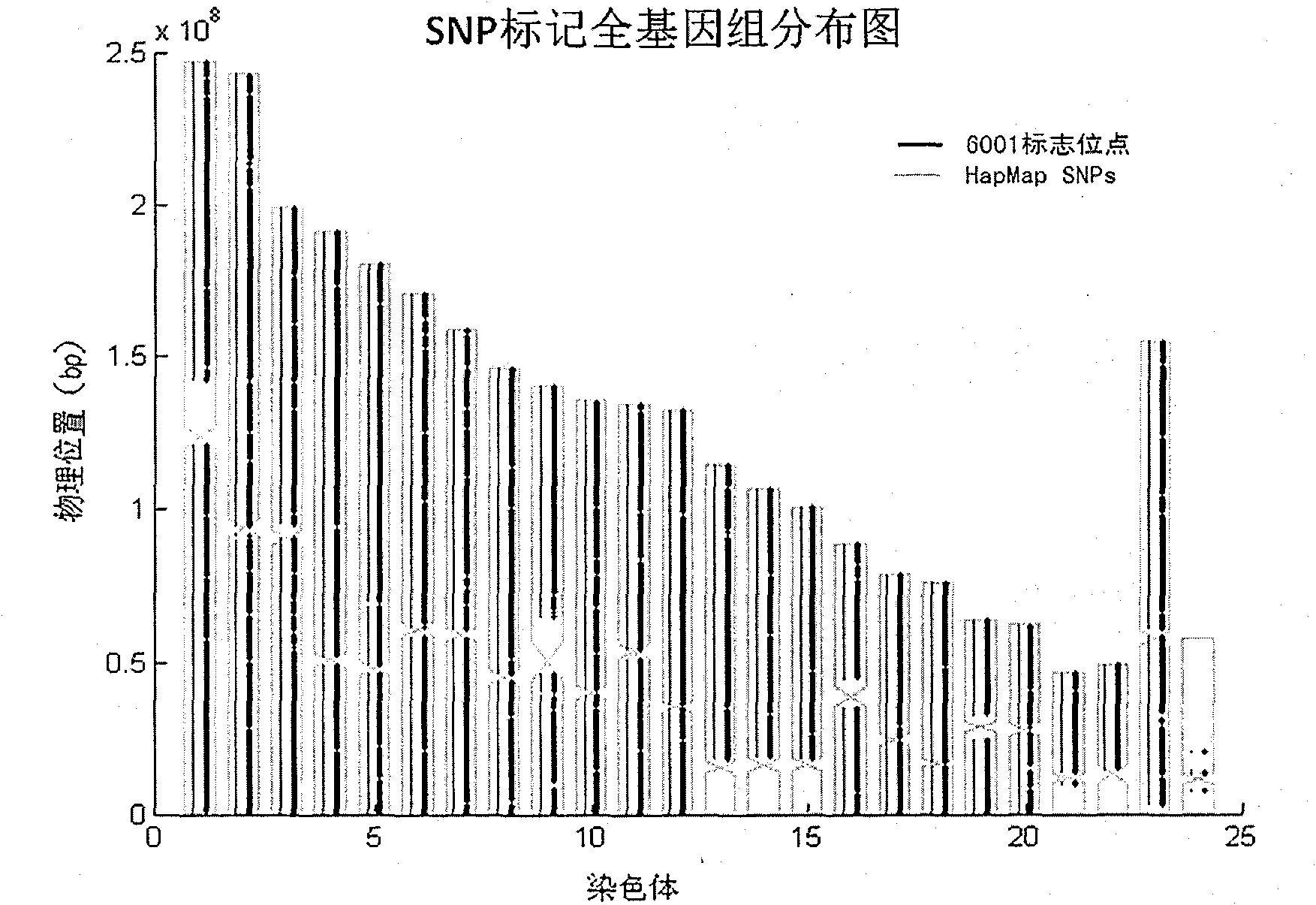
[0038] Probe preparation: Custom synthesis of oligonucleotide probes for all 6001 SNP sites (Illumina typing system). The oligonucleotide probe is a nucleotide sequence containing the SNP site and its upstream and downstream tens of bp, and the SNP site contains two bases that are complementary to the dimorphic base of the SNP. Therefore, corresponding to one SNP site, there are two kinds of single nucleotide probes. Different nucleotide probe preparation companies have slight differences in the processing of probes. Usually, the synthetic oligonucleotide probes with SNP polymorphic sites are immobilized on tiny magnetic beads and attached to special silicon beads. In the microwell of the glass slide, or directly fixed on the slide, a large number of probes can be placed in a very small space, so as to achieve the high-throughput genotyping efficiency of the microchip.
[0039]Collection and storage of family sampl...
Embodiment 2
[0052] Example 2. Linkage Analysis Using Marker Locus Genotypes
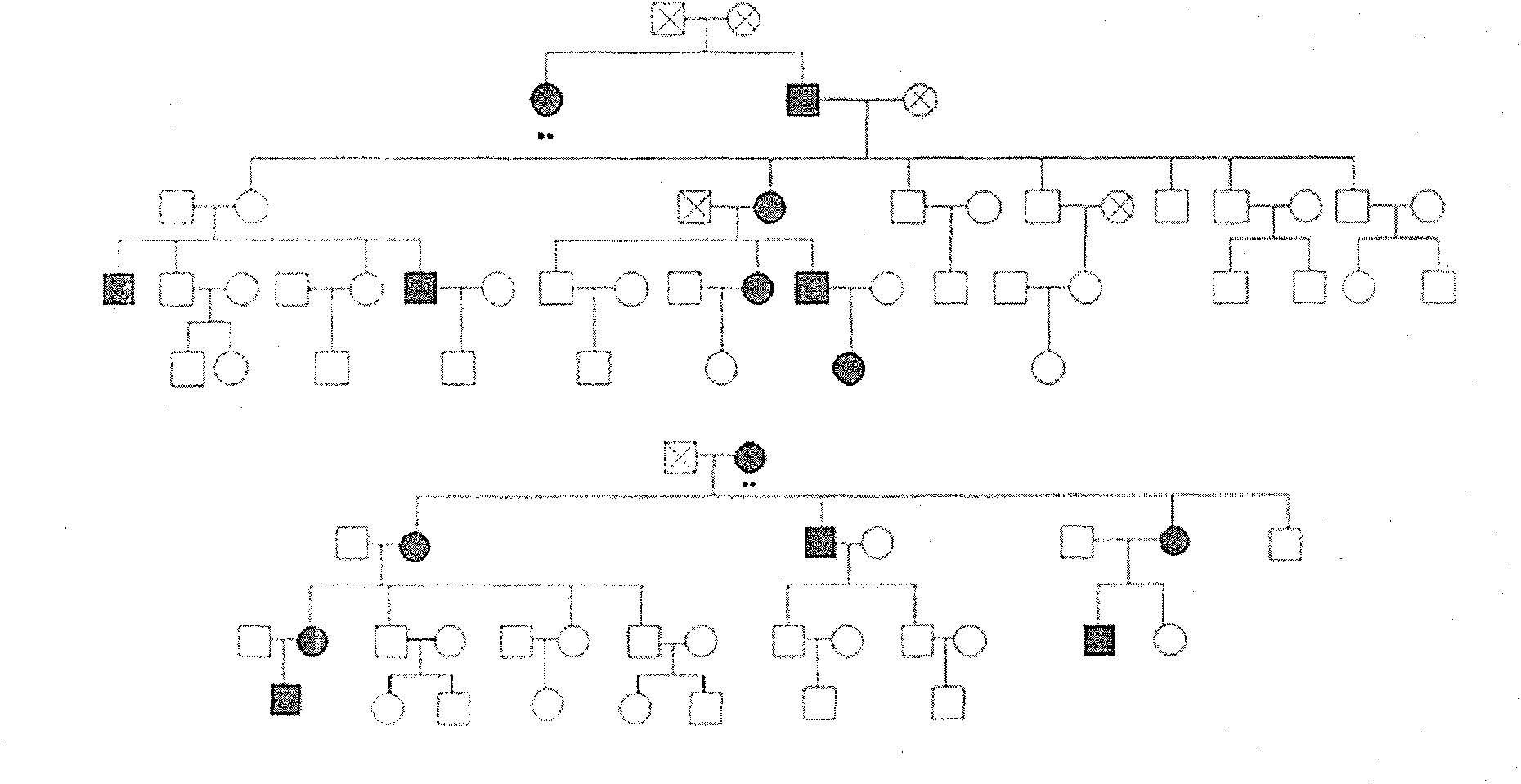
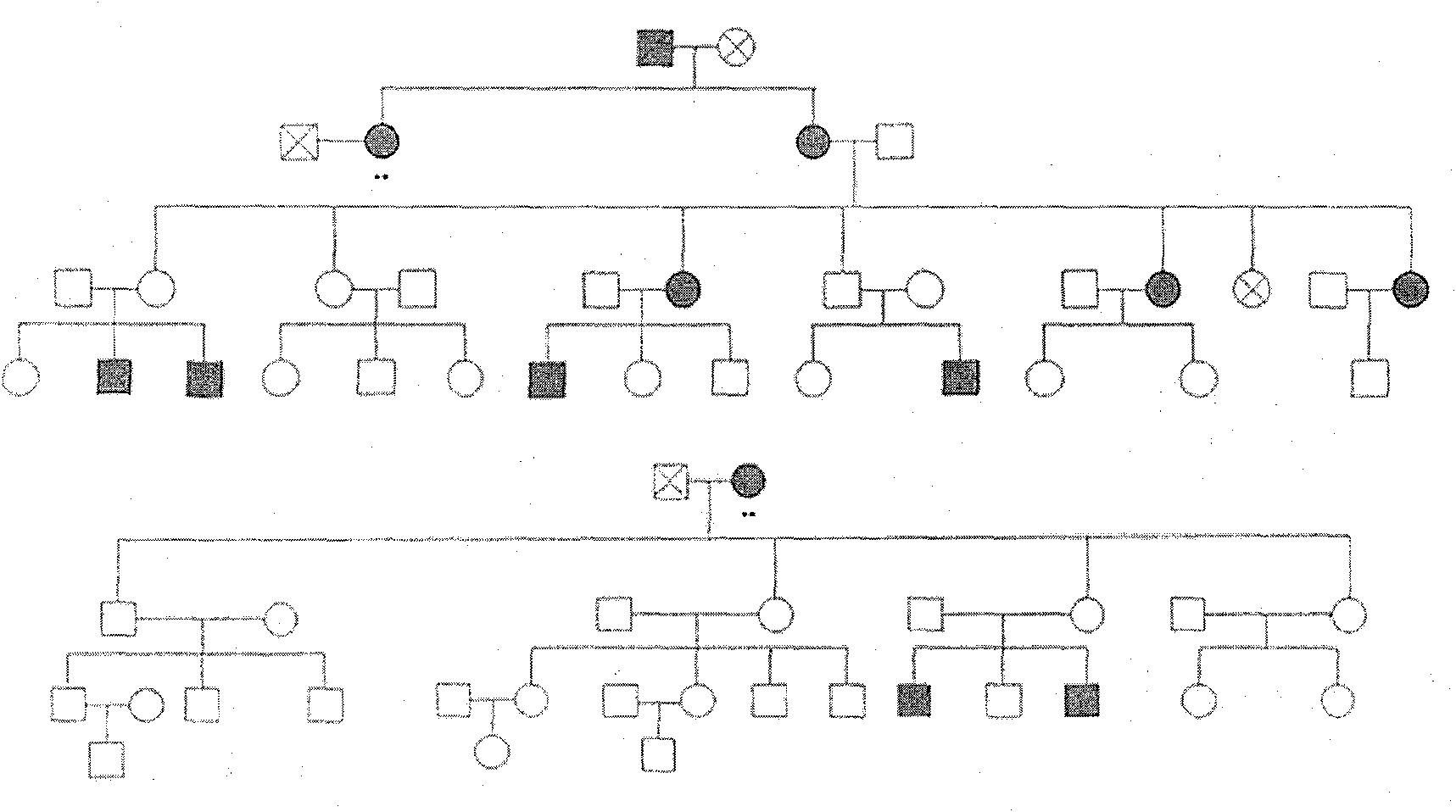
[0053] Localization of pathogenic factors in two families with retinitis pigmentosa.
[0054] Experimental objects: two families as Figure 4 with Figure 5 shown. The first family was from Yongqing County, Hebei Province, including 77 individuals, 14 of whom were sick, and a total of 43 blood samples were taken, 36 of whom entered the experiment and linkage analysis; the second family was in Heze, Shandong, including 59 individuals, of whom 12 people were sick, and blood samples were taken from 23 people, 16 of whom participated in the experiment and analysis process. There was no consanguineous marriage in either family.
[0055] According to the characteristics of the families, the two RP families were determined to be autosomal dominant, and some family members were carriers of disease factors. RP showed incomplete penetrance in both families.
[0056] Preparation of sample DNA: As described in the meth...
Embodiment 3
[0074] Example 3. Effect Verification
[0075] After the disease gene candidate regions were obtained as in Examples 1 and 2, STR marker pairs were selected near the candidate regions to verify the localization results.
[0076]The STR markers D1S425 and D8S1771 were selected in the target region of chromosome 8 and chromosome 1 in family 1 and the experiment was completed. The linkage analysis software Mlink was used to analyze the STR data, and the LOD values were 1.710 and 1.852 respectively; The two candidate regions selected nearby STR markers D14S258 and D14S68 with LOD values of 3.160 and 2.600, respectively. Considering that the selectable STR loci cannot completely fall within the candidate chromosomal region and the heterozygosity of STR itself in the RP family, this result better verifies the linkage relationship between the candidate region and RP disease.
[0077] By searching the gene functions in the candidate region and literature reports, a gene Nrg1 rela...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com