Duolang sheep SNP (single-nucleotide polymorphism) marker, and screening method and application thereof
A screening method, the technology of Duolang sheep, applied in the field of SNP markers and screening of Duolang sheep, can solve the problems of restricting promotion and application, high cost, reducing the accuracy of sheep breed identification, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
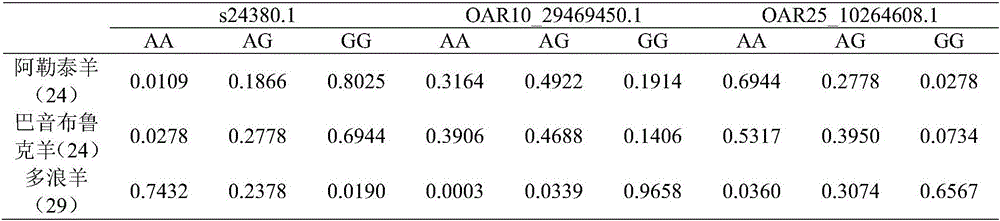
[0036] This embodiment provides a method for screening Duolang sheep SNP markers in Altay sheep and Bayinbulak sheep, including the following steps:
[0037] (1) Select 108 sheep sample individuals including Altay sheep, Bayinbulak sheep and Duolang sheep, collect blood from these sheep, use heparin sodium for anticoagulation, and use Tiangen blood genome kit to extract genomic DNA, 0.8% agarose Gel electrophoresis detection, nucleic acid protein quantifier to determine its concentration and purity, dilute to 100ng / ul, and save for later use.
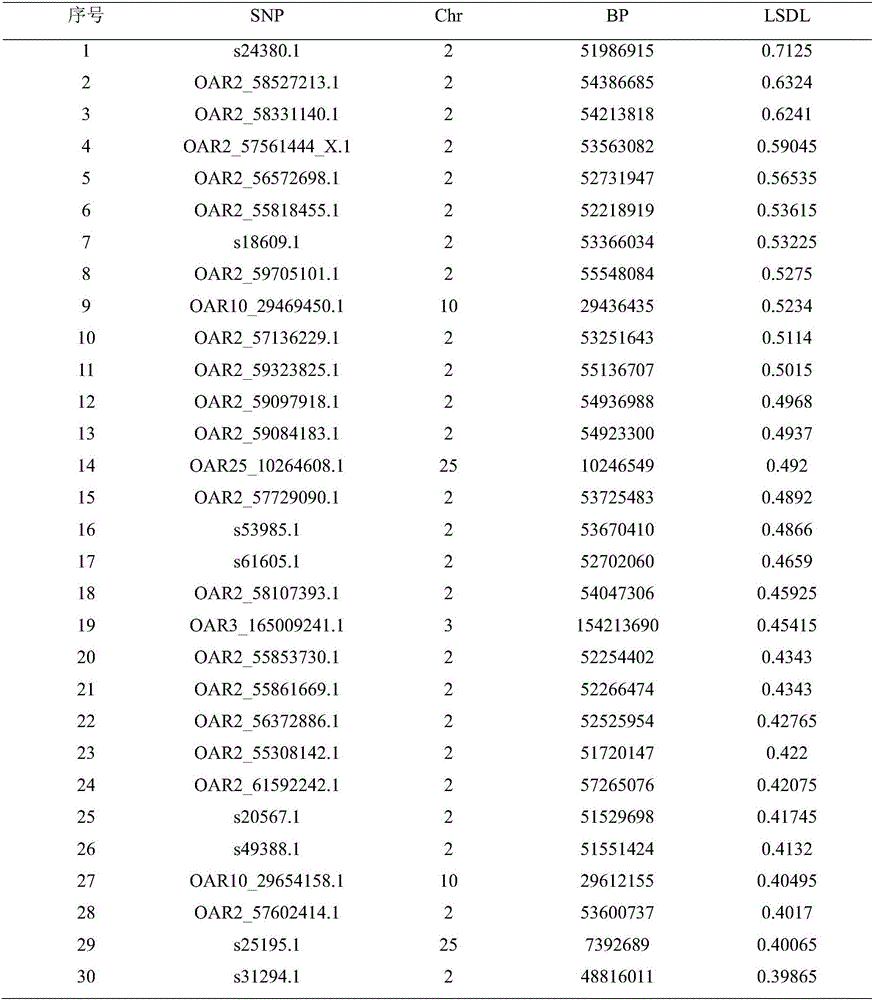
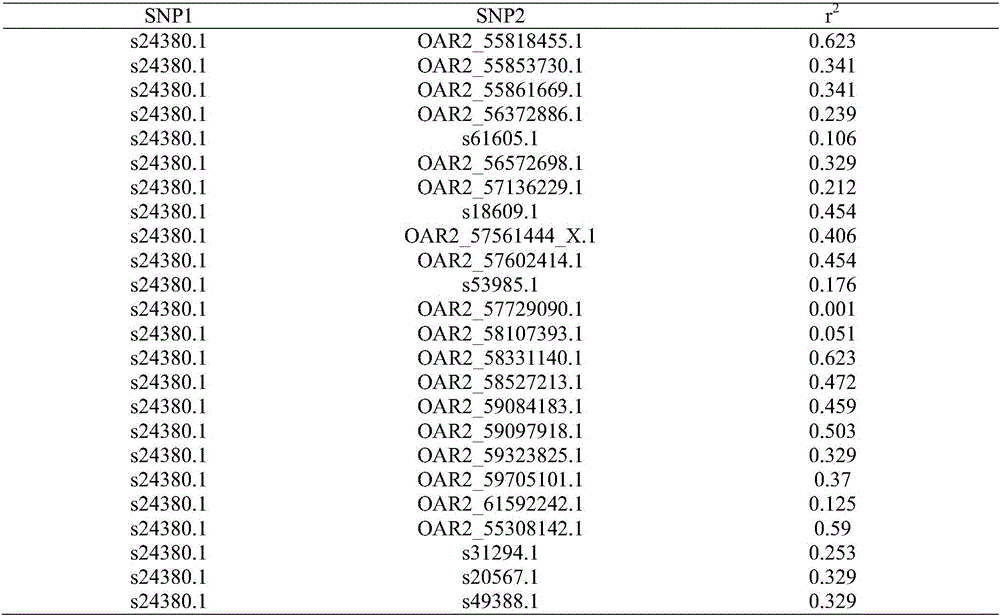
[0038] (2) Perform the genotype analysis of the prepared DNA samples on the sheep whole genome 50K SNP chip (Illumina Ovine 50KBeadchip), and perform quality control on the 108 individuals and 54 241 SNPs obtained with Plink software: individual detection rate (call rate)> 0.9, SNP detection rate> 0.9, minimum allele frequency (MAF)> 0.01, Harberger balance (HWE)> 0.000001. Since the detection rate of one sheep is less than 0.9, after quali...
Embodiment 2
[0065] This embodiment provides a method for screening Duolang sheep SNP markers in German meat merino and Chinese merino sheep, including the following steps:
[0066] (1) Select 159 individual sheep samples containing German meat merino, Chinese merino sheep and Duolang sheep, collect the blood of these sheep, use heparin sodium for anticoagulation, and use Tiangen blood genome kit to extract genomic DNA , 0.8% agarose gel electrophoresis detection, nucleic acid protein quantifier to determine its concentration and purity, dilute to 100ng / ul, save for later use.
[0067] (2) Perform genotype analysis of the prepared DNA samples on the sheep whole genome 50K SNP chip (Illumina Ovine 50KBeadchip), and perform quality control on 54 241 SNPs of 159 individuals with Plink software: individual detection rate (call rate)> 0.9, SNP detection rate> 0.9, minimum allele frequency (MAF)> 0.01, Harberger balance (HWE)> 0.000001. After quality control, 60 German meat merino (GMM), 60 Chinese ...
Embodiment 3
[0090] The comparison between the identification method of the present invention and the existing method for identifying Duolang sheep is shown in Table 14.
[0091] Table 14 Comparison of the identification method of the present invention and the existing method of identifying Duolang sheep
[0092]
[0093] The Duolang sheep SNP marker of the present invention can be applied to the identification of Duolang sheep breeds or their livestock products. When the Duolang sheep SNP marker of the present invention is used to identify Duolang sheep breeds or their livestock products, it overcomes The defects of phenotypic identification are simple and easy to implement, and there is no interference between sites, which greatly improves the accuracy of the identification of Duolang sheep breeds or their livestock products and effectively reduces costs.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com