Method for detecting single nucleotide polymorphism of genes related to yean trait of milk goat
A technology of single nucleotide polymorphism and detection method, which is applied to SNPs molecular markers and application fields related to lambing traits of dairy goats, and can solve the problems of reducing regional heterozygosity and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0017] Materials and experimental methods used in the present invention:
[0018] 1. Experimental materials
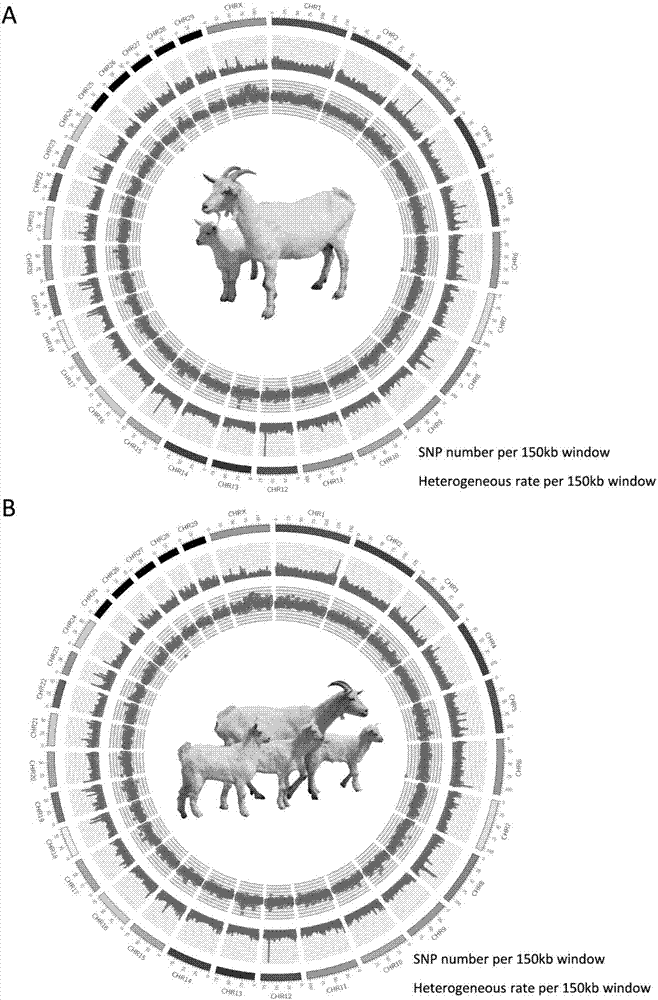
[0019] 37 Laoshan dairy goats, samples were collected from Qingdao Aote Special Sheep Farm. Two groups of Laoshan dairy goats with large differences in lambing performance. In the low-yield group, 20 dairy goats gave birth to 1 lamb, and in the high-yield group, there were 13 lambs. The lambing number of an individual is 3, and the lambing number of a dairy goat is 4. The sample was collected from dairy goat ear muscle tissue.
[0020] 2. DNA extraction and sequencing of dairy goat ear muscle tissue
[0021] The DNA of 37 dairy goats was extracted using the QIAamp DNA Mini Kit (QIAGEN, 51304, Hilden, Germany). The extracted DNA was mixed in equal amounts for library construction, and then the high-yield and low-yield populations were subjected to high-throughput sequencing using the Illumina HiSeq 2000 sequencing platform.
[0022] 3. Data Analysis
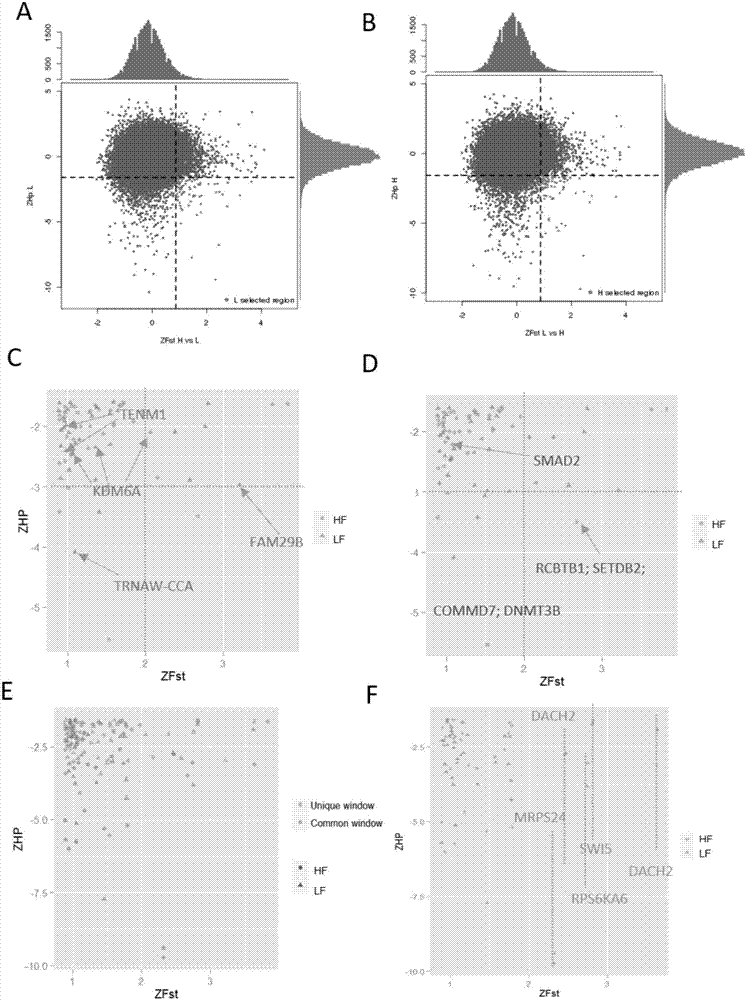
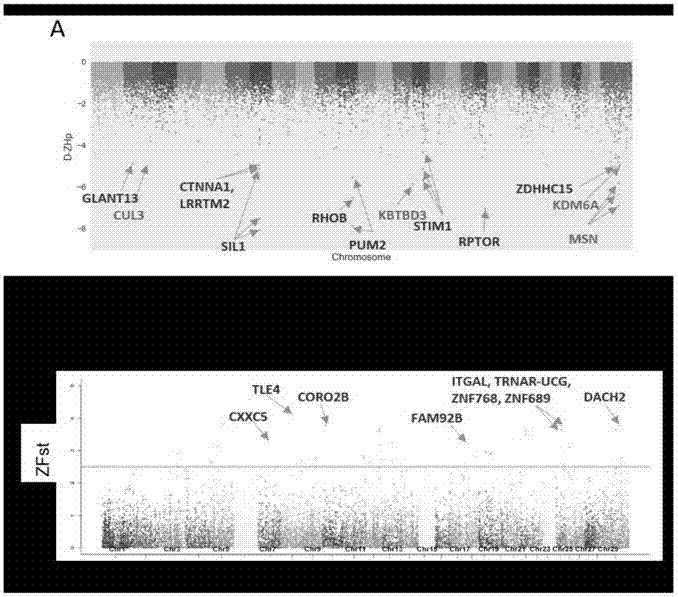
[0023] 3.1 Se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com