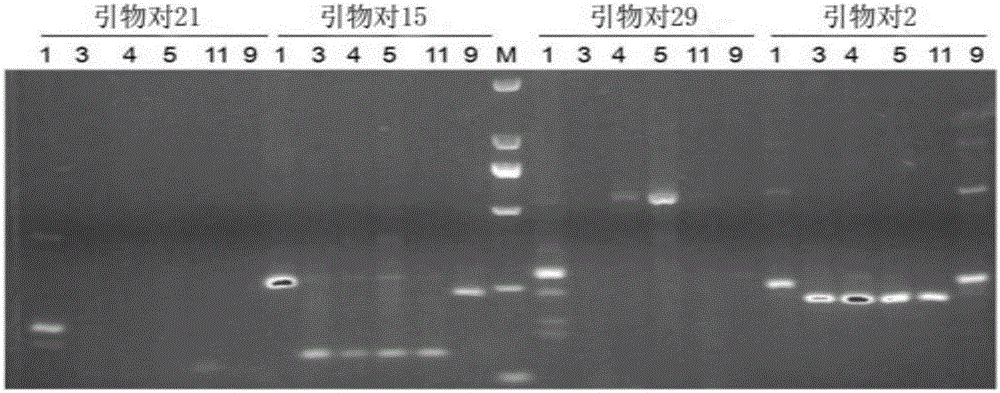
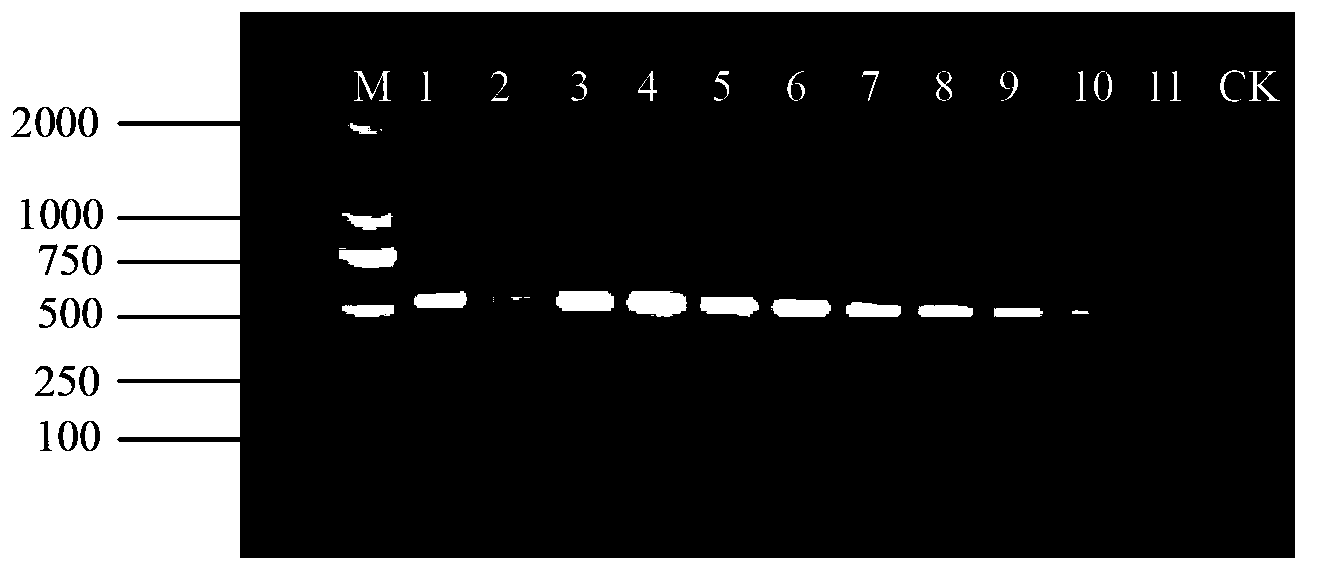Patents
Literature
59 results about "Sibling species" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Sibling species are species that are very similar in appearance, in behavior and in other characteristics, but they are reproductively isolated. In other words, sibling species are pairs or groups of genetically closely related species which are often morphologically indistinguishable, but are reproductively isolated, meaning that while they may interbreed, the offspring cannot reproduce. Sibling species may arise as a result of allopatric speciation through geographic isolation, parapatric speciation, or sympatric speciation. The important thing to note is that sibling species are in fact separate biological species. The biological species concept, proposed by Ernst Mayr in 1942, emphasizes reproductive isolation as the basis of defining a species. The definition states: "A species is defined as a population or group of populations whose members have the potential to interbreed with one another in nature and to produce viable offspring, but cannot produce viable, fertile offspring with members of other species."
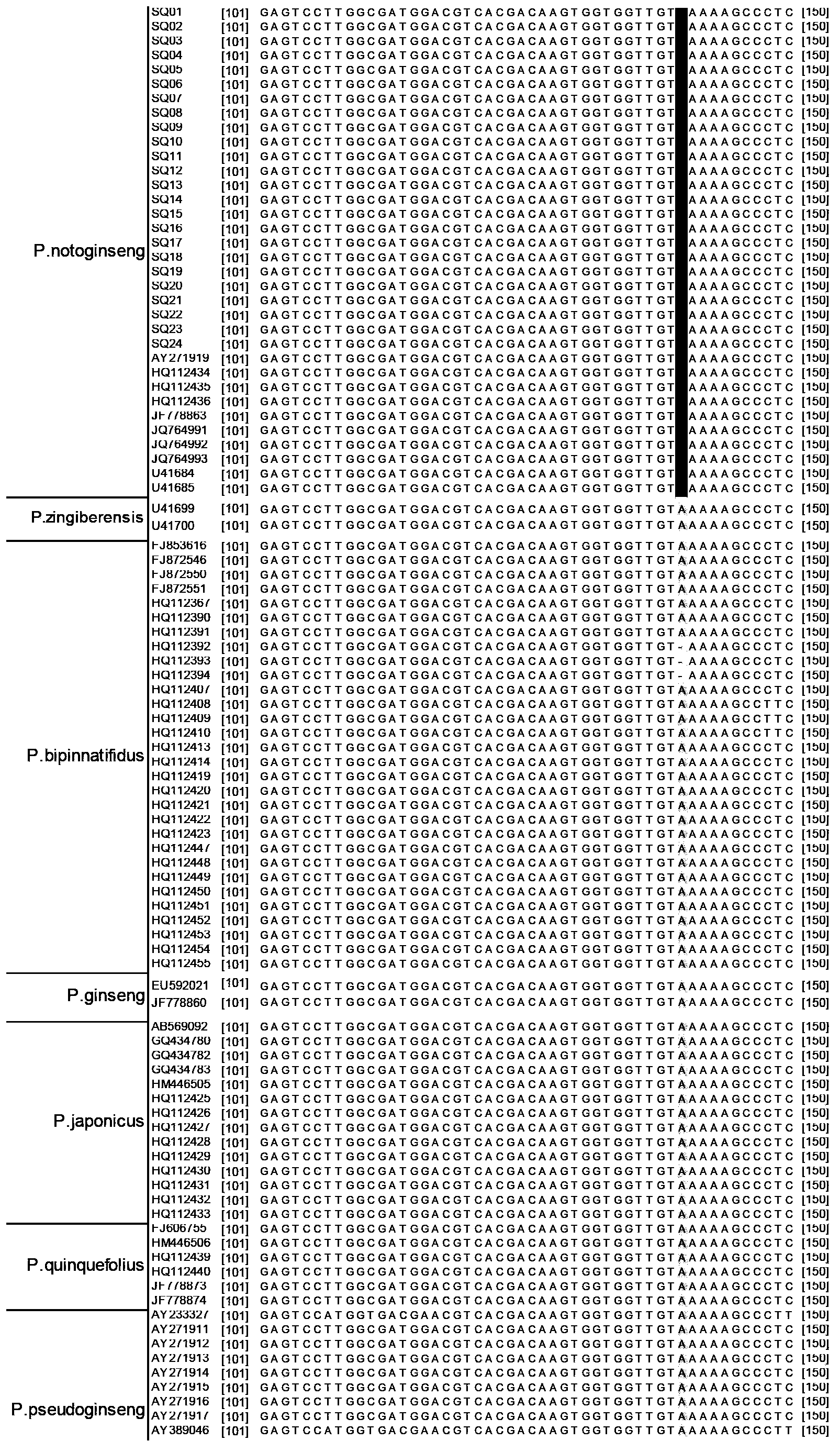
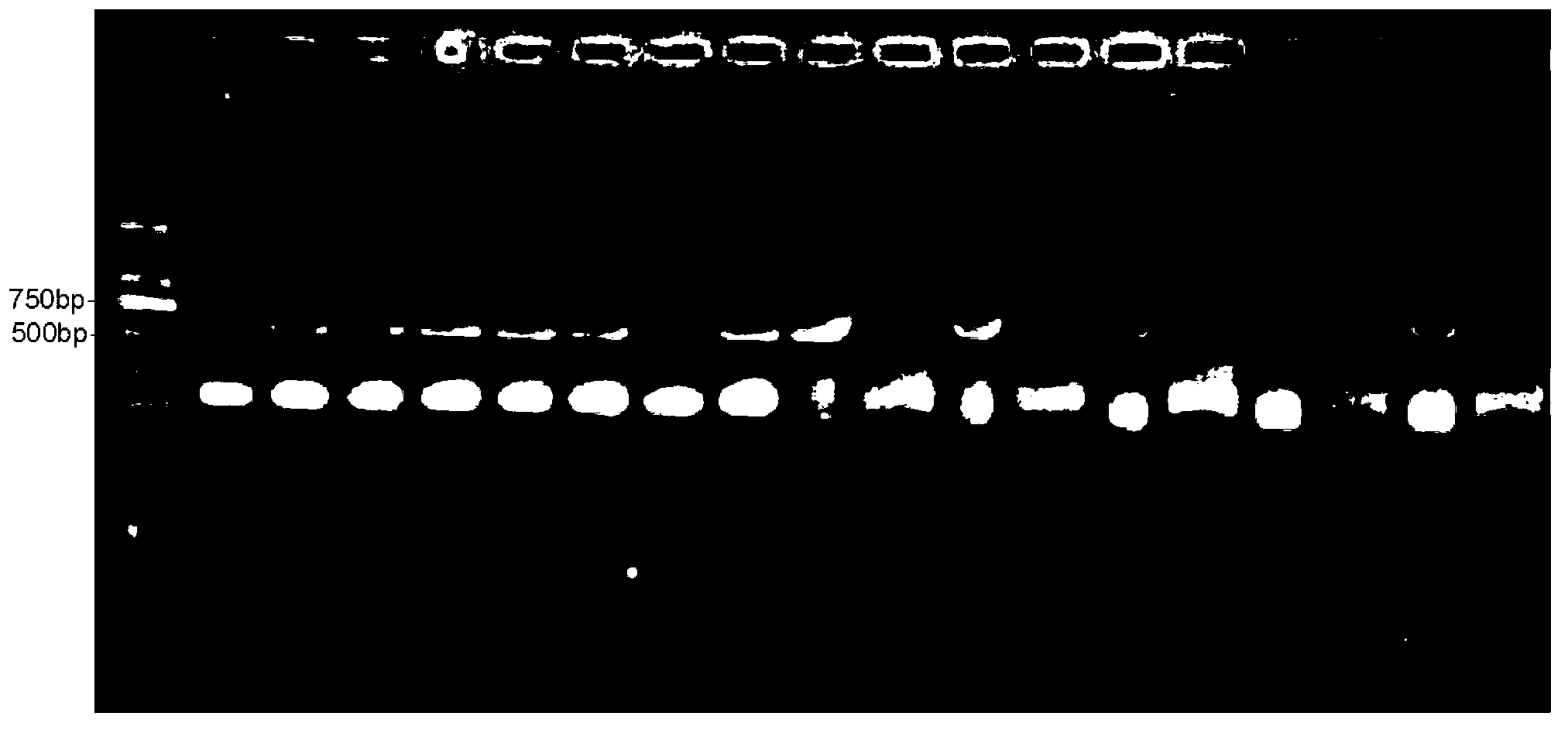
Method for quickly identifying pseudo-ginseng
InactiveCN102888456ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentation3-deoxyriboseAdulterant
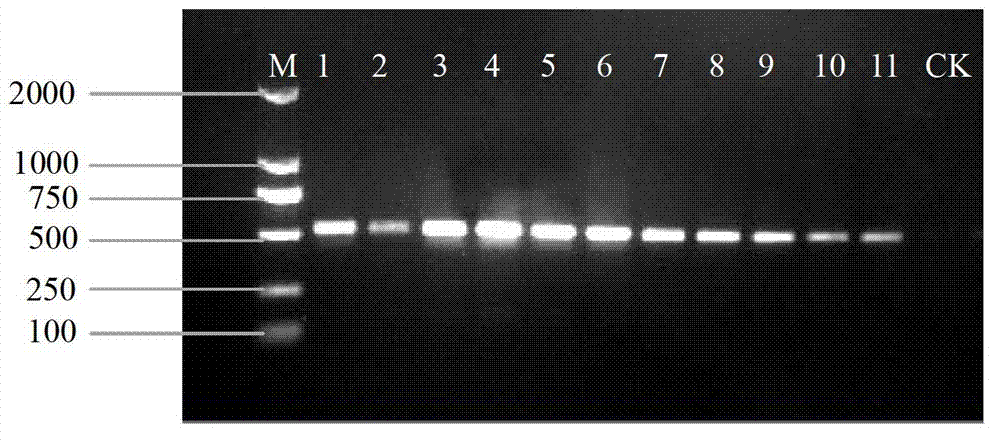
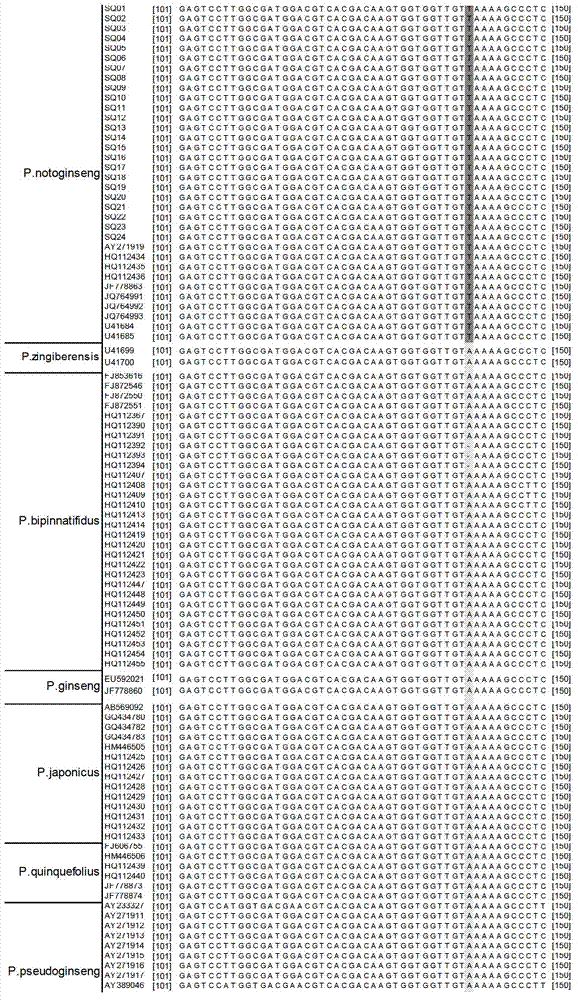
The invention discloses a method for quickly identifying pseudo-ginseng. The method comprises steps as follows: 1) getting DNA (Deoxyribose Nucleic Acid) of a sample to be detected as a template, and amplifying the fragments containing the sequences shown as SEQ ID No.1 (Sequence Identification Number); and 2) sequencing the amplified products, analyzing the sequences as shown in SEQ ID No.1; if the bit 140 beginning from 5' end in the sequences as shown in SEQ ID No.1 is T or the bit 207 is T, identifying that the sample is pseudo-ginseng; and if the bit 140 and the bit 207 beginning from the 5' end in the sequence as shown in SEQ ID No.1 are not T, identifying that the sample is not pseudo-ginseng. With the adoption of the method disclosed by the invention, the raw pseudo-ginseng, the sibling species of the raw pseudo-ginseng, the crude medicine, the raw medicinal slices, the powder and the adulterant can be quickly and accurately identified.
Owner:INST OF MEDICINAL PLANT DEV CHINESE ACADEMY OF MEDICAL SCI
Forage grass maize production process
InactiveCN1926962AIncrease productionStrong stress resistancePlant genotype modificationHorticultureRelationship - FatherSibling species
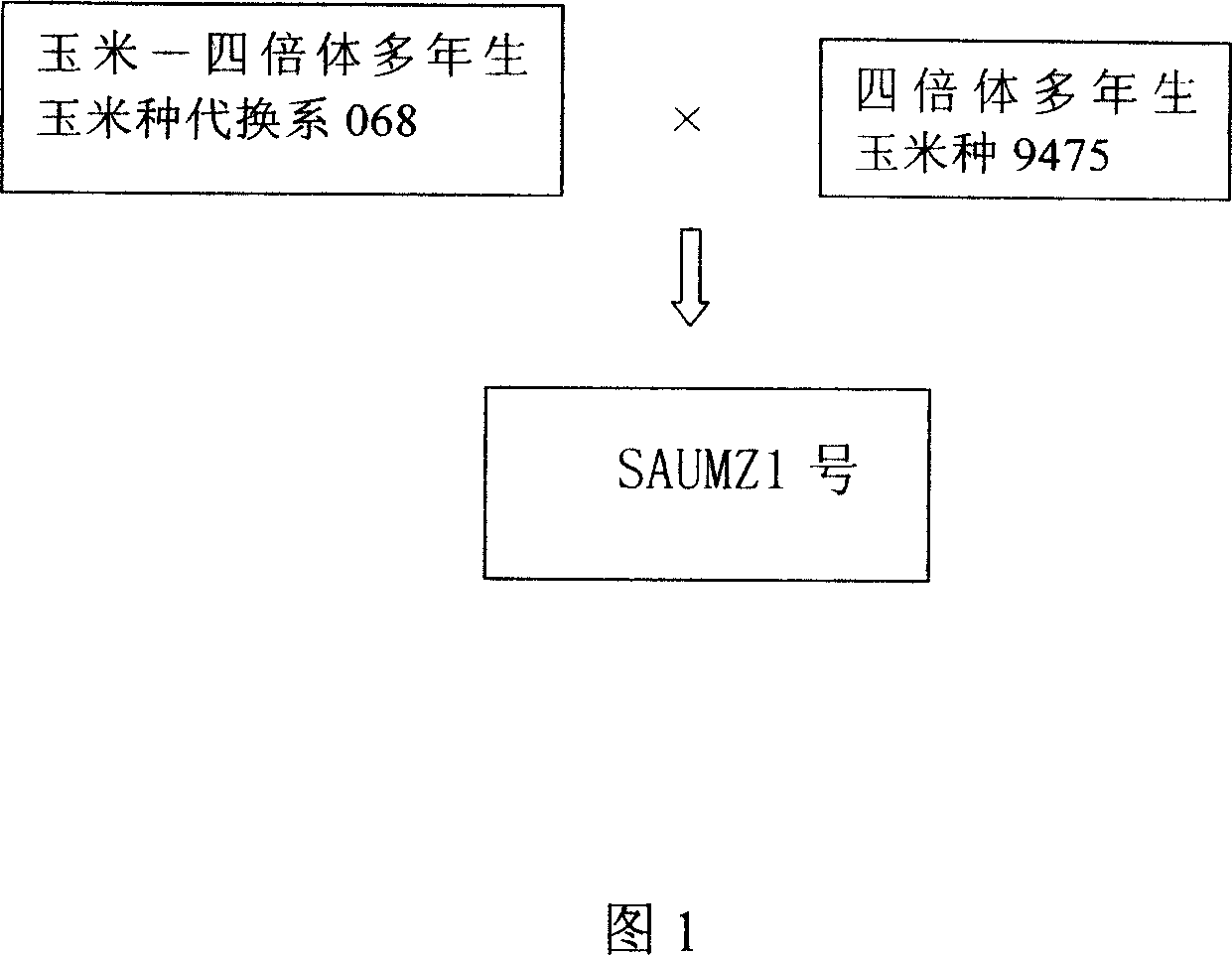
The invention relates to a method for producing forage grass corn hybrid. Wherein, it uses the corn-tetraploid corn substitution system 068 as mother, uses the corn sibling species tetraploid corn 9475 as father to cross to obtain the forage grass corn SAUMZ1. And it adjusts the seeding time, cuts capillament, adjusts the planting density, emasculates from the mother, adjusts the flower period, pollinates artificially, and harvests at right time. The invention can improve yield and quality.
Owner:SICHUAN AGRI UNIV
Process for selecting and breeding forage grass maize strain by using maize sibling species
InactiveCN1926963ASolve the problem of low seed setting rate of hybridizationIncreased biomassPlant genotype modificationSibling speciesForage
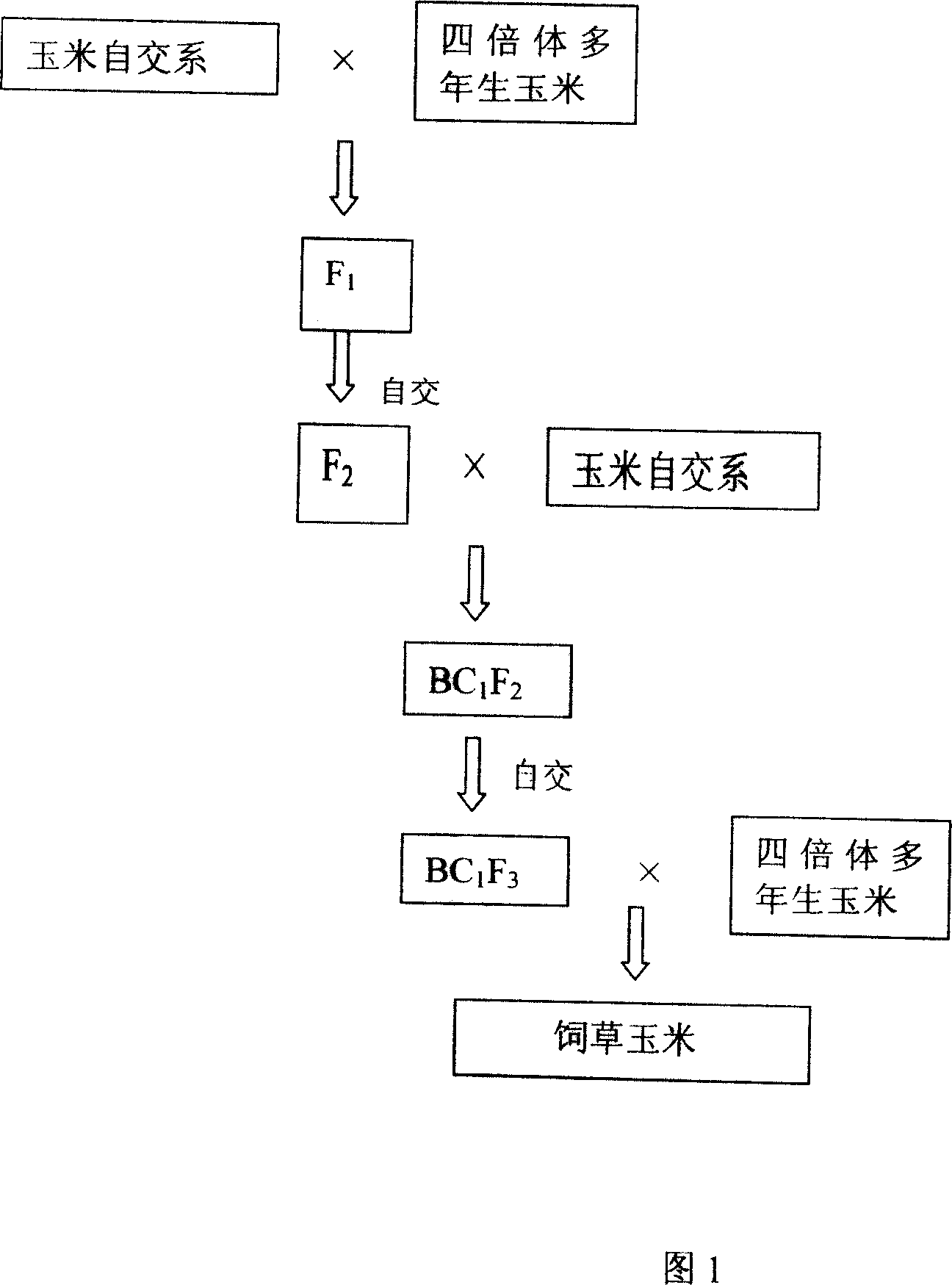
The invention relates to a method for method for using corn to breed forge grass corn kind. Wherein, it uses corn autocopulation system and tetraploid grown to cross and backcross to obtain corn-tetraploid grown corn substitution system; then crosses replace system and tetraploid grown corn to obtain the forge grass corn, which has high yield and high quality.
Owner:SICHUAN AGRI UNIV
Specific primer based kit for identifying five common cryptoleste stored grain insects
ActiveCN103952463ASimplify molecular detection stepsSimple and fast operationMicrobiological testing/measurementAgricultural scienceCryptolestes
The invention discloses a specific primer based kit for identifying five common cryptoleste stored grain insects. The kit provided by the invention comprises at least one pair of the following 5 pairs of primers: a primer pair 1 (sequence 1 and sequence 2), a primer pair 2 (sequence 3 and sequence 4), a primer pair 3 (sequence 5 and sequence 6), a primer pair 4 (sequence 7 and sequence 8) and a primer pair 5 (sequence 9 and sequence 10). Experiments show that compared with a traditional morphological identification method, the cryptoleste stored grain insect species identification method using the kit provided by the invention does not need morphological taxonomy base and realizes the identification of non-adult state; andcompared with other molecular biological identification methods, the method simplifies the molecular detection steps for identification of common cryptoleste stored grain insects, has the characteristics of simple operation, short time consumption, high sensitivity, stability and visual results. The invention lays a foundation for further research on identification of sibling species of cryptoleste.
Owner:CHINA AGRI UNIV
Breeding method of alien substitution line
The invention relates to a breeding method of an alien substitution line, which can be used for further shifting exogenous beneficial genes to wheat by utilizing a wheat alien substitution line so that an alien translocation line is bred. The alien substitution line has the very important function at the aspects of gene mapping of wheat sibling species, researches on the hereditary feature of an exogenous chromosome in the background of wheat and the evolutionary relationship between the wheat and the sibling species thereof and the like, and a wheat variety with abundant hereditary basis and excellent comprehensive character is selected as a receptor material for breeding the alien substitution line so that the utilization value of the alien substitution line can be enhanced. In addition, a technology system for conveniently and rapidly breeding the alien substitution line is established and also is one of important research subject for enhancing the breeding efficiency of the alien substitution line.
Owner:李祥
SS-COI (Sirex noctilio F. specific cytochrome oxidase I) primer pair and quick molecular detection method
InactiveCN104232634AReduce the need for technical expertiseThe identification method is fast and stableMicrobiological testing/measurementDNA/RNA fragmentationTremex fuscicornisSibling species
Owner:BEIJING FORESTRY UNIVERSITY
Rapid molecular identification method for cinnamon
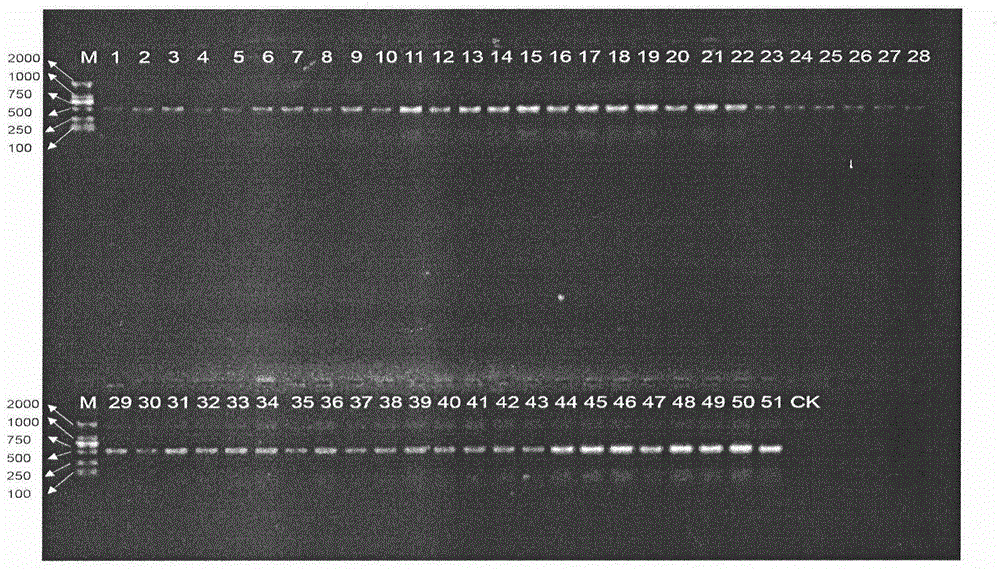
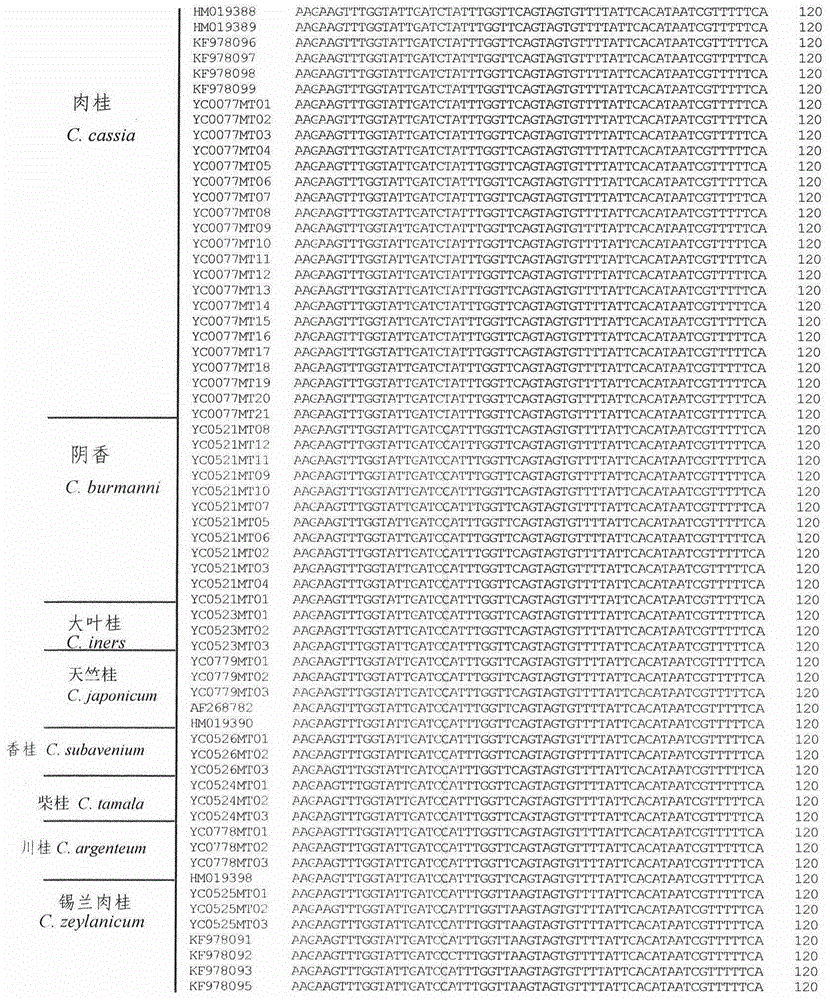
InactiveCN104611445ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationAdulterant
The invention discloses a rapid cinnamon identification method which comprises the following steps: (1) by taking to-be-detected sample DNA as a template, amplifying a sequence shown as SEQ ID No.1; (2) sequencing the amplification product, analyzing the sequence shown as SEQ ID No.1, identifying the product to be cinnamon if the 80th site from the 5' end of the sequence shown as SEQ ID No.1 is T, and identifying the product to be not cinnamon if the 80th site from the 5' end of the sequence shown as SEQ ID No.1 is C. According to the method disclosed by the invention, the original cinnamon plants and sibling species thereof, medicinal materials, raw medicinal slices, powder and adulterants can be rapidly and accurately identified.
Owner:INST OF MEDICINAL PLANT DEV CHINESE ACADEMY OF MEDICAL SCI
Persimmon tree colletotrichum gloeosporioide SSR (Simple Sequence Repeat) primer pair developed based on sibling species genome and application of primer pair
ActiveCN106701943AEffective prevention and controlMicrobiological testing/measurementMicroorganism based processesConsanguinityGenome
The invention belongs to the technical field of biologics, and in particular discloses a persimmon tree colletotrichum gloeosporioide SSR (Simple Sequence Repeat) primer pair developed based on sibling species genome and application of the primer pair. According to the persimmon tree colletotrichum gloeosporioide SSR primer pair, 30 pairs of SSR primers are developed by finding SSR by using MISA software on the basis of the genome of a sibling species, namely, colletotrichum gloeosporioides, of persimmon tree colletotrichum gloeosporioides, and 15 pairs of polymorphism SSR primers are obtained by screening 6 different persimmon tree colletotrichum gloeosporioides; the persimmon tree colletotrichum gloeosporioide SSR primer pairs provided by the invention are novel markers which exist stably, can be used for analyzing genetic diversity and genetic differentiation on the persimmon tree colletotrichum gloeosporioides, can be also used for identifying varieties of the persimmon tree colletotrichum gloeosporioides and studying ties of consanguinity, and have very important significances in effective prevention and control on the persimmon tree colletotrichum gloeosporioides.
Owner:河南省林业科学研究院
Determination method for HPLC specific chromatogram of codonopsis tubulosa kom dried root
The invention discloses a determination method for the HPLC specific chromatogram of codonopsis tubulosa kom dried root. According to the present invention, codonopsis tubulosa kom dried roots from more than 10 different production places are collected, and are respectively prepared into samples, the HPLC maps and the UV spectrograms of the samples and a reference substance lobetyolin are obtained, and after the lobetyolin peak is determined as the reference peak, the HPLC reference specific chromatogram of the codonopsis tubulosa kom dried root is constructed by using a traditional Chinese medicine chromatographic fingerprint similarity evaluation system so as to provide the basis for the effective identification of the codonopsis tubulosa kom dried root from other codonopsis pilosula herbs having the sibling species and provide a set of feasible and effective scheme for the quality control of the codonopsis tubulosa kom dried root; and the determination method has characteristics of simpleness, easy performing, and good use effect.
Owner:GUIYANG COLLEGE OF TRADITIONAL CHINESE MEDICINE
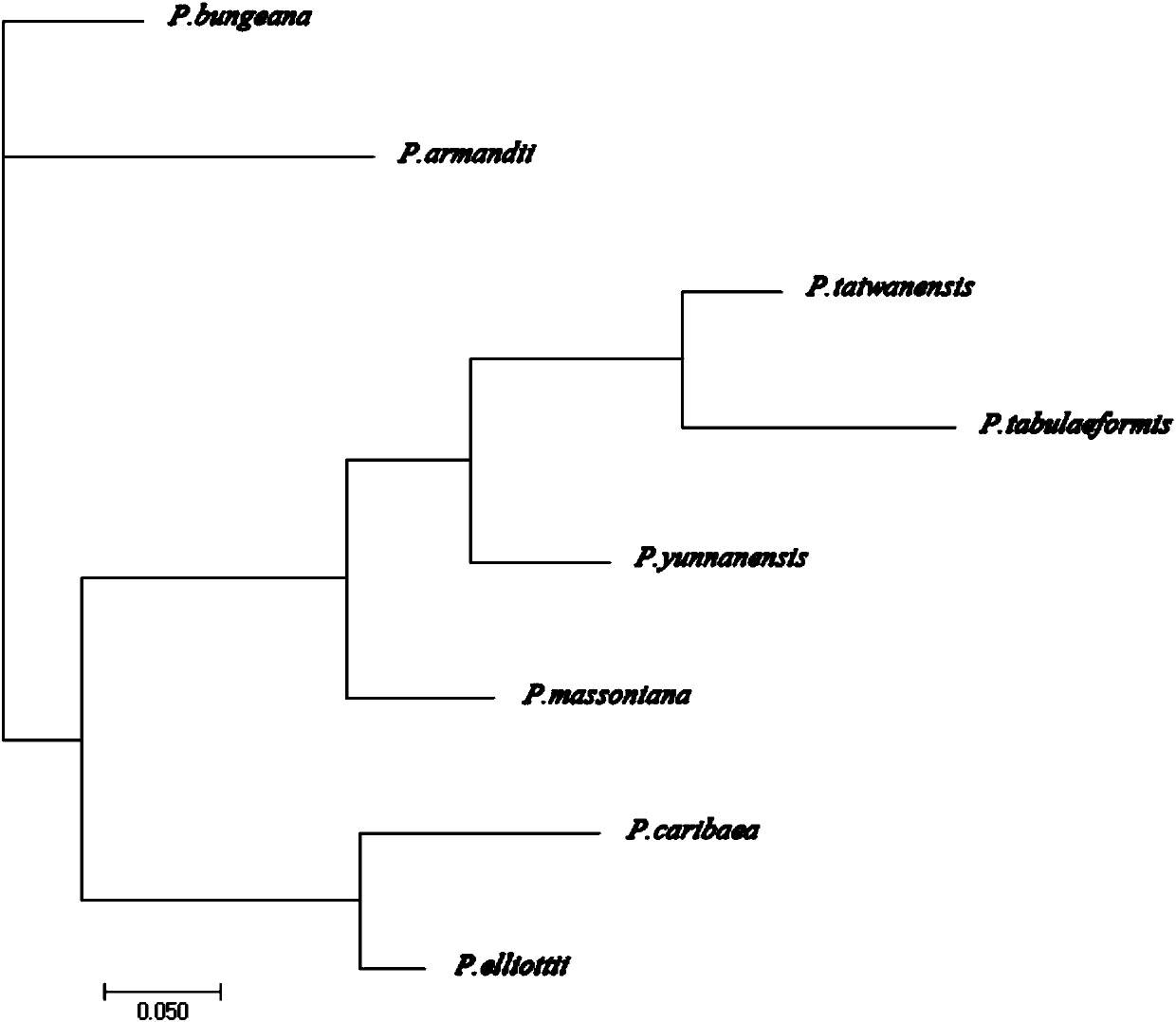
Pinus massoniana cpSSR polymorphism primer and identification method of pine sibling species of primer
ActiveCN107557362ALess prone to recombinationMultiple copies smallMicrobiological testing/measurementDNA/RNA fragmentationCapillary electrophoresisPhylogenetic tree
The invention discloses a pinus massoniana cpSSR polymorphism primer and an identification method of pine sibling species of the primer. According to design of a chloroplast genome of pinus massoniana, 16 pairs of cpSSR primers are obtained, further, 7 pairs of primers which are stable in difference and good in polymorphism are screened from the 16 pairs of cpSSR primers, and through utilization of the 7 pairs of primers, a finger print and phylogenetic tree of 8 kinds of pine sibling species are successfully established. Based on the 7 pairs of cpSSR primers, a PCR amplification reaction is carried out. Capillary electrophoresis technology is used to analyze a PCR product, 8 kinds of pine sibling species trees can be accurately identified. The primer and method are efficient, accurate, and convenient and are not prone to be affected by the environment. The polymorphism primer and an identification method thereof can reflect the genetic information of the 8 kinds of pine plasmons, andcan be used for research of identification of 8 kinds of pine sibling species and genetic relationship.
Owner:NANJING FORESTRY UNIV
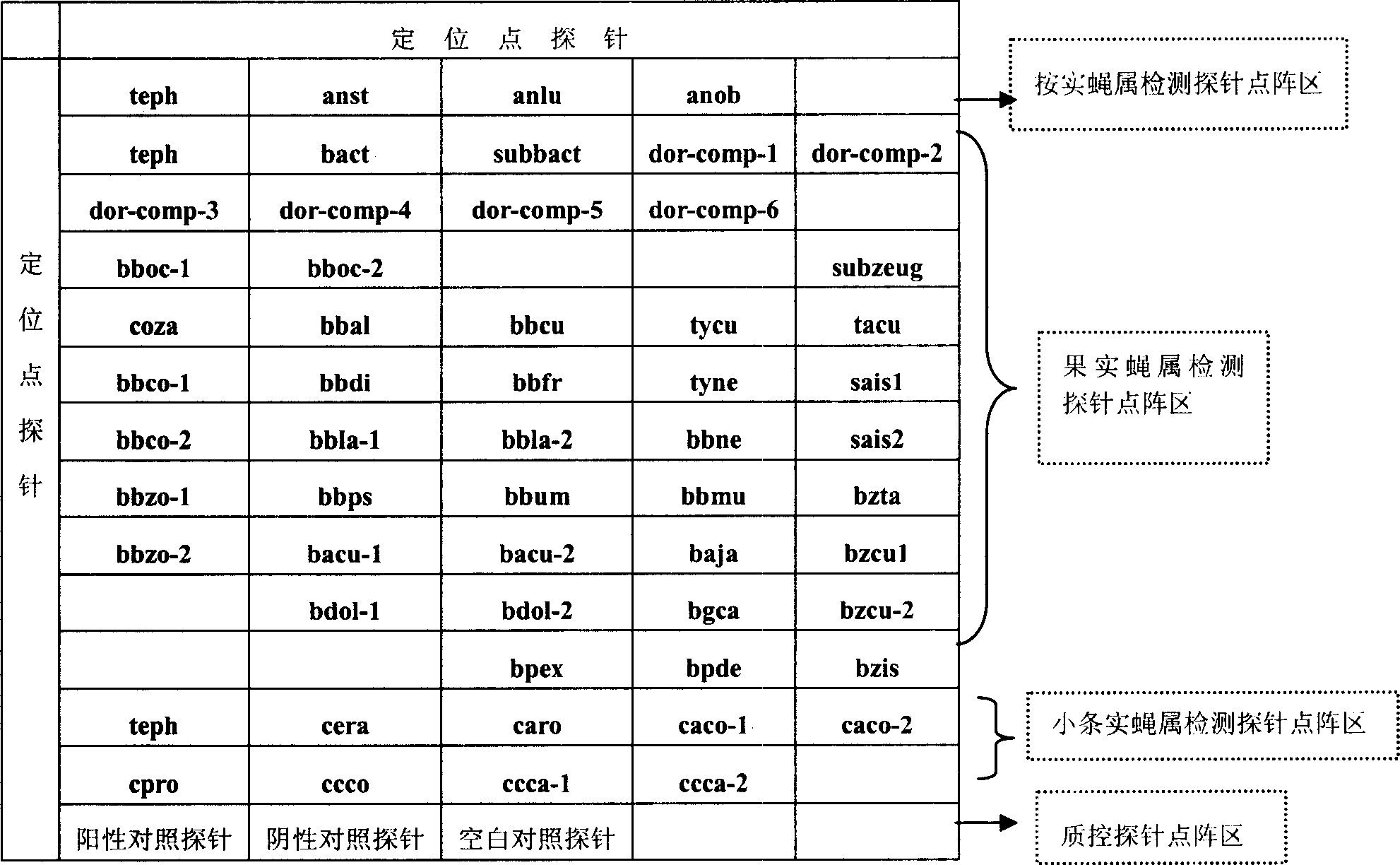
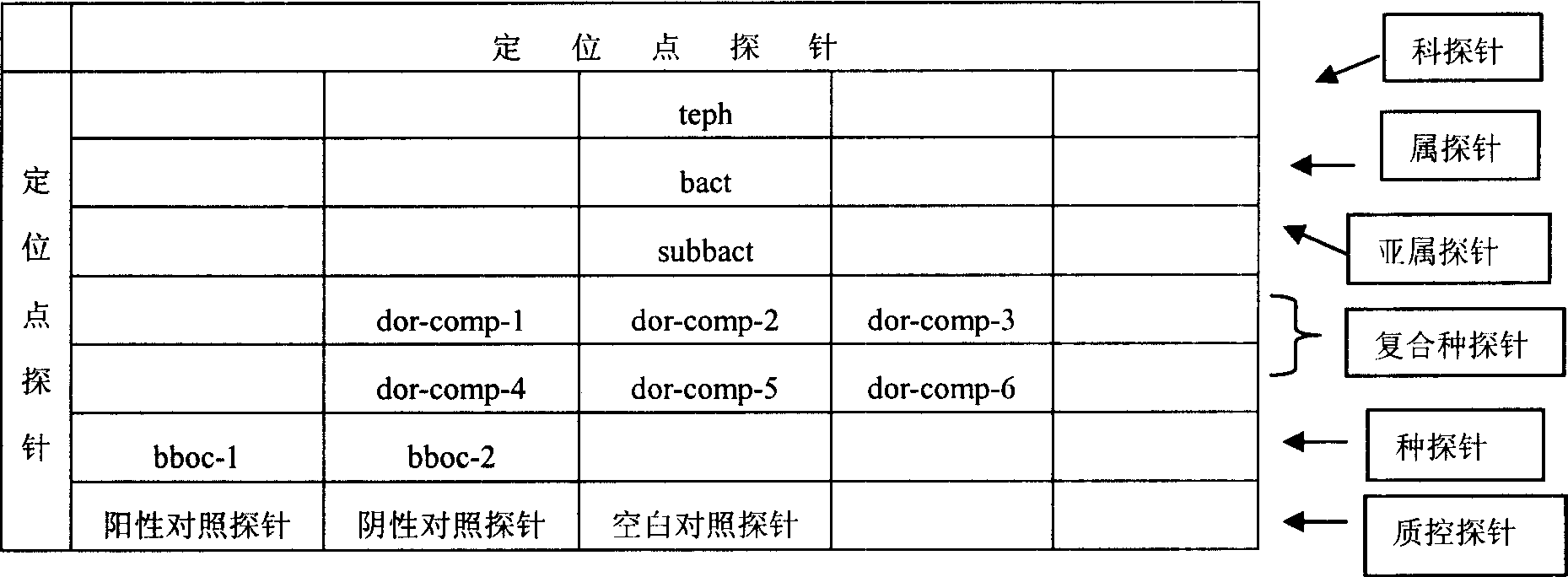
Fruit-fly classified detection biochip, detection method and reagent kit
InactiveCN1995393ARapid identificationAccurate identificationMicrobiological testing/measurementFluorescence/phosphorescenceTephritidaeGeneral purpose
The invention discloses a biological chip, testing method and testing agent box to classify Tephritidae, which selects probe of general-purpose probe and atopic probe or subgeneric general purposed probe and composite group / sibling species atopic probe to identify Tephritidae rapidly and precisely.
Owner:SHENZHEN AUDAQUE DATA TECH
Fingerprint spectrum of cauliflower hybrid and construction method thereof
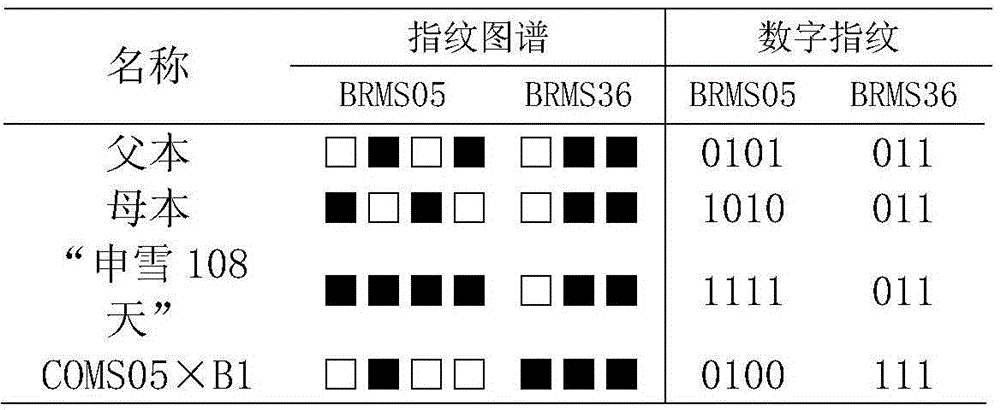
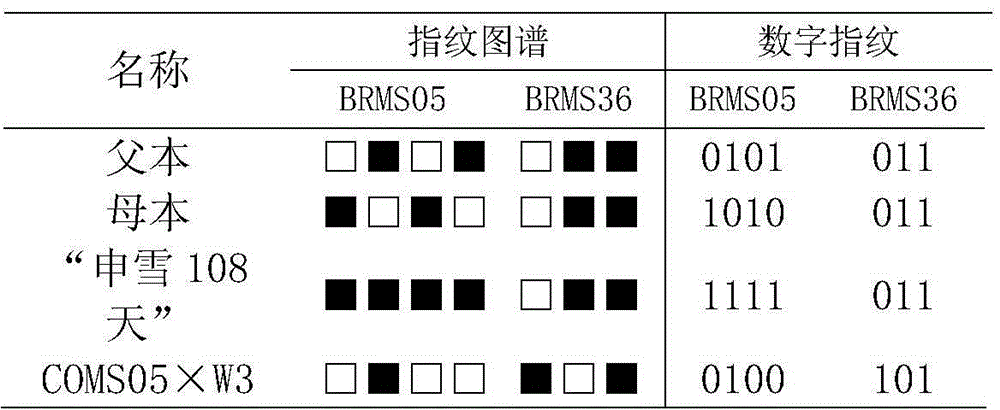
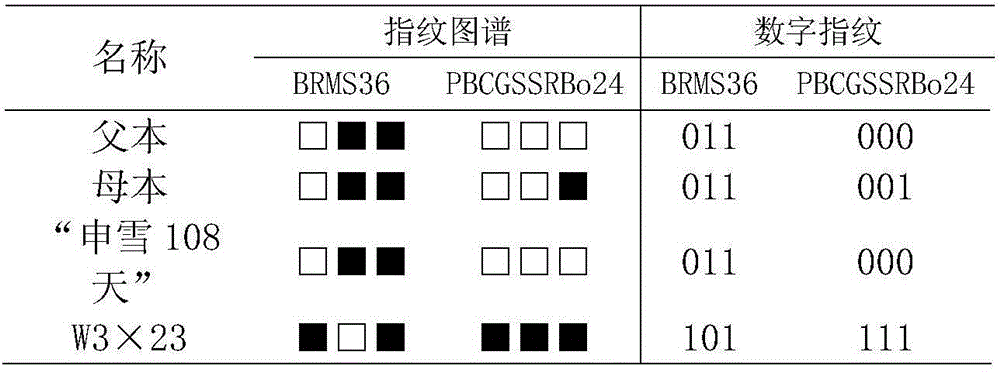
InactiveCN104561338AUniqueness guaranteedPrevent counterfeitingMicrobiological testing/measurementOrganismSibling species
The invention belongs to the technical field of molecular organisms and particularly relates to a fingerprint spectrum of cauliflower hybrid and a construction method thereof. The cauliflower hybrid is temporarily named as 'shenxue 108 day', and is a new cauliflower variety bred by multi-year hybrid in Shanghai funong seed industry limited company. The fingerprint spectrum and the construction method have the advantages that SSR molecular markers are utilized for carrying out quantitative marking on the 'shenxue 108 day', and the fingerprint spectrum different from sibling species or the same series of cauliflower varieties is constructed; the applicability to identification, selling and truth-falseness discrimination of farmers for the shenxue 108 day of the cauliflower is achieved. The fingerprint spectrum and the construction method have the effects that the mastering is easy and the benefit for producers and seed buyers can be protected.
Owner:SHANGHAI JIAO TONG UNIV
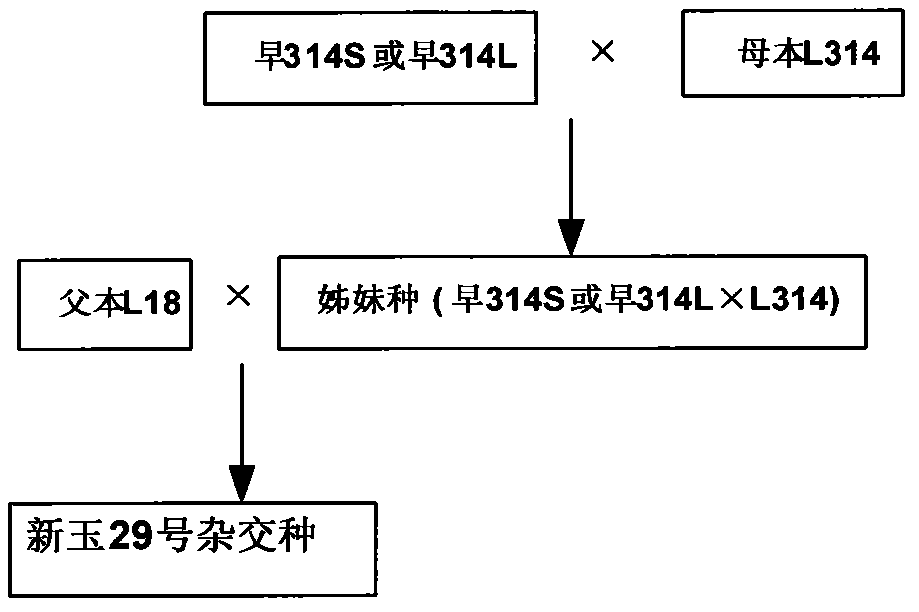
High-yield seed production method of early-ripening maizes
InactiveCN103168675AReduce manufacturing costImprove seed productionPlant genotype modificationHybrid seedAgricultural science
The invention discloses a high-yield seed production method of early-ripening maizes. The near relation sister line 314 S or early 314 L is crossbred with the original female parent L314 to generate a sibling species as the female parent, and the L18 is used as the male parent to be prepared into a hybrid species, i.e., Xinyu NO. 29, so that the seed production yield is improved, and the production cost of seeds is further reduced; meanwhile, compared with the original Xinyu NO. 29, the Xinyu NO. 29 prepared by the sister line hybrid seeds retains the consistency in yield, stability, adaptability and agronomical character; 30 mu of the Xinyu NO. 29 produced with the sibling species are planted in Company 5, Southern District, 163rd Regiment of 9th Agricultural Division, Tacheng District, Xinjiang Jiuhe Seed Company in 2008, the average seed production yield is 376.6 kg / mu, and the high yield reaches 442.6 kg / mu, and is increased by 40 percent than the original hybrid species; and 1500 mu of the Xinyu NO. 29 produced with the sibling species are planted in 164th Regiment, Tacheng District in 2009, the seed production yield is 460 kg / mu, and through the seed production of the sibling species, the seed production yield is effectively improved, and the cost is reduced.
Owner:INST OF GRAIN CROPS XINJIANG ACAD OF AGRICLTURE SCI
Primer pair for identifying enteromorpha sibling species, DNA (Deoxyribonucleic Acid) bar code, and application and detection methods thereof
ActiveCN107988408ABreak through overdependenceEfficient use ofMicrobiological testing/measurementDNA/RNA fragmentationDNA barcodingA-DNA
The invention belongs to the technical field of alga molecular markers, and concretely relates to a primer pair for identifying enteromorpha sibling species, a DNA (Deoxyribonucleic Acid) bar code, and application and detection methods thereof. The primer pair for identifying the enteromorpha sibling species comprises: an upstream primer sequence: 5'GCTGACATACCATCACGATAGT 3'(SEQ ID U01); a downstream primer sequence: 5'CGTATTATTTCACCAGACCCTT 3'(SEQ ID U02). The invention relates to application of the primer pair in identifying the enteromorpha sibling species. The primer pair for identifying the enteromorpha sibling species, the DNA bar code, and the application and detection methods thereof provided by the invention are strong supplement for traditional species identification, automationand standardization can be realized during a sample identification process, the heavy dependency on experiences is broken, fragments of algae can be utilized for being quickly and effectively identified, and an application system easy to use can be built within a shorter time.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Detection kit for aeromonas veronii and application method thereof
InactiveCN106399520AStrong specificityQuick checkMicrobiological testing/measurementMicroorganism based processesDuplex pcrSibling species
The invention provides a detection kit for aeromonas veronii and an application method thereof. In the technical scheme, gyrB gene is found to be a single-copy house-keeping gene through experiments and has more remarkable advantages than conventional 16S rRNA in the distinguishing and identifying of aeromonas veronii and sibling species thereof; and meanwhile, Aha gene is found to be stably exist in the aeromonas veronii and related to the toxicity thereof. Based on the beneficial discoveries, the gyrB gene and Aha gene are adopted as target genes to develop a duplex PCR detection method for aeromonas veronii; in the method, specific primers are designed for the two target genes, and a PCR reaction system and reaction conditions are determined. In the invention, the sensitivity and specificity of aeromonas veronii detection are high, the pelteobagrus fulvidraco-source pathogenic aeromonas veronii can be quickly and accurately detected, and the wrong detection and missed detection are effectively avoided. Meanwhile, repeated culture and redundant series of biochemical reactions are prevented, and the time, labor and cost are saved.
Owner:SOUTHWEST UNIVERSITY
Whole genome DNA of scylla paramamosain mitochondria and testing method
InactiveCN103060332AFast detection methodEfficient detectionMicrobiological testing/measurementGenetic engineeringScylla paramamosainGermplasm
The invention relates to a whole genome DNA of scylla paramamosain mitochondria and a testing method. The DNA molecule which is in a closed ring structure has a length of 15824 bp; and the testing method comprises the steps of: (1) collecting relative gene sequences of the scylla paramamosain mitochondria and comparing the relative gene sequences of the scylla paramamosain mitochondria with whole genome sequences of mitochondria of sibling species; (2) designing a primer and carrying out PCR (Polymerase Chain Reaction) amplification; and (3) sequencing PCR products and assembling the sequences. According to the invention, the whole genome DNA sequences of the scylla paramamosain mitochondria are obtained. The testing method disclosed by the invention has the advantages of being fast in speed, low in cost, easy to master and the like; and the obtained whole genome DNA of the mitochondria can be applied to the fields such as heritable variation analysis of the scylla paramamosain, genetic resource authorization and protection, phylogeny and the like.
Owner:EAST CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Operation mode of species quarantine and identification center
InactiveCN106520518AResolve inconsistenciesSolve the inconvenient preservation of specimensBioreactor/fermenter combinationsBiological substance pretreatmentsDNA barcodingVisual inspection
The invention discloses an operation mode of a species quarantine and identification center. The operation mode comprises the following steps: issuing of a sample acquisition tool, a sample acquisition pipe and a transporter, a sample acquisition information record sheet and a species identification system client to a first port, sample acquisition, pretreatment of sample inspection, sample presentation, sample receiving, acquisition of sample physical information, detection of sample DNA barcode and result feedback. By the establishment of a novel medical vector DNA barcode quarantine and identification mode and standardized processes of sampling, sample presentation, detection, etc., fast and accurate identification of imported medical vectors is realized, and the problem that detection of medical vectors, especially vector insects is limited by development condition and limb mutilation, sibling species are hard to accurately identify, some female insects cannot be identified and imported medical vectors cannot be accurately and quickly identified due to lack of reference data and reference specimens is solved.
Owner:INSPECTION AND QUARANTINE TECHNOLOGY CENTER ZHONGSHAN ENTRY EXIT INSPECTION AND QUARANTINE
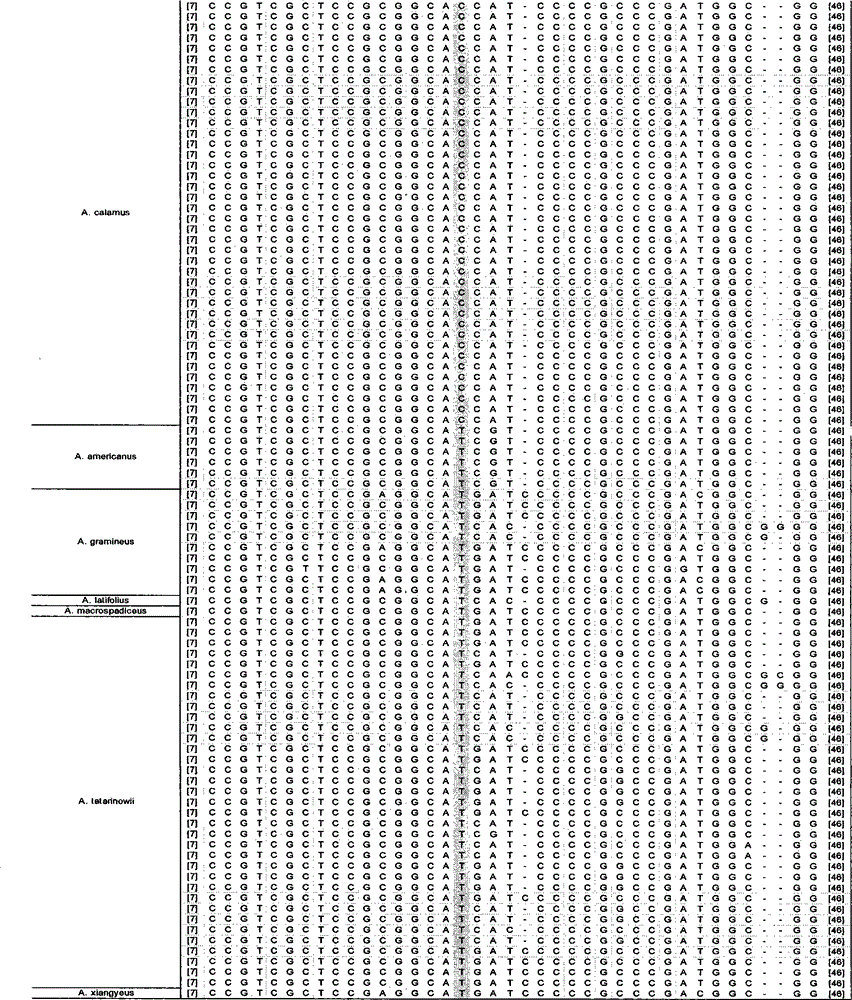
Rapid identification method of Rhizoma Acori Calami
InactiveCN104480105ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceAcorus calamus
The invention discloses a rapid identification method of Rhizoma Acori Calami. The method comprises the following steps: 1) DNA of a sample to be tested is used as a template, and fragments containing sequence as shown in SEQ ID No.1 are amplified; and 2) amplification products are sequenced, and the sequence as shown in SEQ ID No.1 is analyzed; if the 23rd site from 5' end of the sequence as shown in SEQ ID No.1 is C and / or the 212th site is G, the sample is identified as Rhizoma Acori Calami; and if the 23rd site from 5' end of the sequence as shown in SEQ ID No.1 is T and the 212th site is C or T, the sample to be identified is not Rhizoma Acori Calami. By the above method, rapid and accurate identification of the Rhizoma Acori Calami orginal plant and its sibling species, medicinal materials, raw medicinal slices and adulterants can be realized.
Owner:INST OF MEDICINAL PLANT DEV CHINESE ACADEMY OF MEDICAL SCI
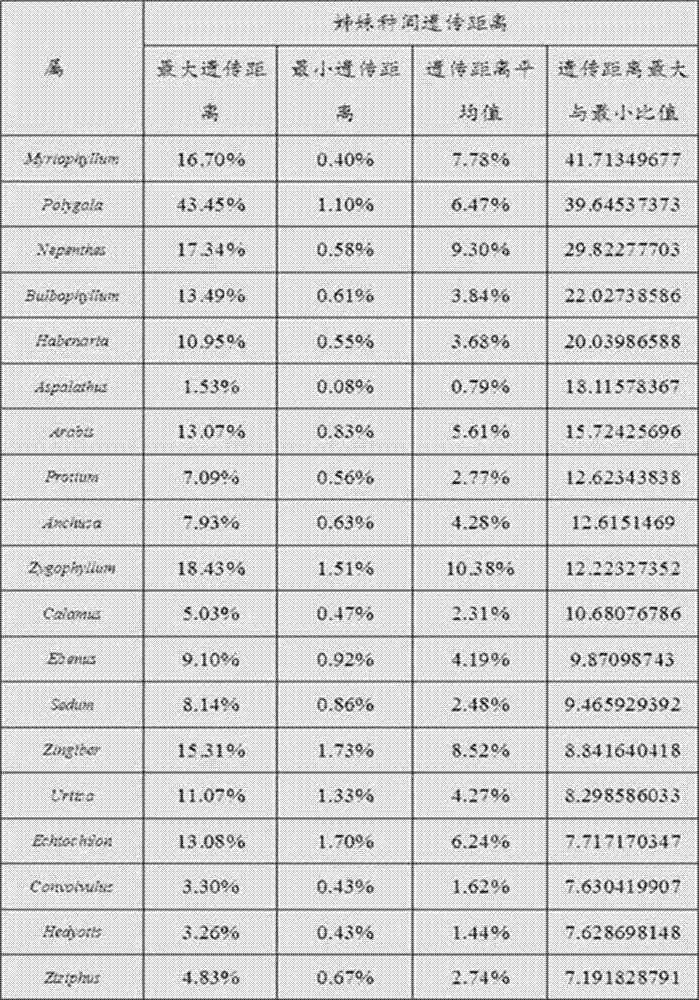
Identification method for seed plant species and application
InactiveCN107400723AImprove accuracyLow coverage requirementsMicrobiological testing/measurementSequence analysisTest sampleSignificant difference
The invention relates to an identification method for seed plant species and application. The identification method comprises the following steps that the seed plant species are selected, ITS2 gene sequences of the seed plant species are obtained, an evolutionary tree is built based on the ITS2 gene sequences, the multiple pairs of sibling species are selected in the evolutionary tree, the genetic distance between each pair of sibling species is calculated, difference significance testing is conducted on the multiple groups of genetic distances of the species belonging to the same phylum, the genetic distances with non-significant differences are selected, the average value of the genetic distances with the non-significant differences is calculated, and thus the molecule critical value is obtained; and the genetic distance between the to-be-tested species and the target species is calculated, and the genetic distance and the molecule critical value of the phylum of the to-be-tested species are compared, so that the species category is determined. According to the identification method for the seed plant species, only ramification degree comparison needs to be conducted between to-be-tested samples or the suspected specifies and the ITS2 gene sequences of the target species, simpleness, convenience and easy operation are better achieved, accuracy is high, objectivity is high, the situation that the large number of other species in the same genus are sampled as a reference is not needed, and the high application value is achieved.
Owner:SHANDONG UNIV
Deoxyribonucleic acid (DNA) bar code standard sequences of sibling species of culicoides latreille, and molecular identification method of sibling species of culicoides latreile
InactiveCN108034733AGuaranteed accuracyEnsure reliabilityMicrobiological testing/measurementFermentationMolecular identificationDNA barcoding
The invention discloses deoxyribonucleic acid (DNA) bar code standard sequences of sibling species of culicoides latreille, and a molecular identification method of the sibling species of the culicoides latreille. The molecular identification method is characterized in that a molecular biology DNA sequencing method is adopted to obtain COI gene sequences of the sibling species, such as culicoideslongirostris, culicoides liui and culicoides albifascia tokunaga, of the culicoides latreille, and then comparison of the similarity of the gene sequences is performed, so that the sibling species ofthe culicoides latreille can be identified according to comparison results. The molecular identification method of the sibling species, such as the culicoides longirostris, the culicoides liui and theculicoides albifascia tokunaga, of the culicoides latreille is beneficial to implementation of rapid and accurate identification of the culicoides longirostris, the culicoides liui and the culicoidesalbifascia tokunaga.
Owner:ZUNYI MEDICAL UNIVERSITY
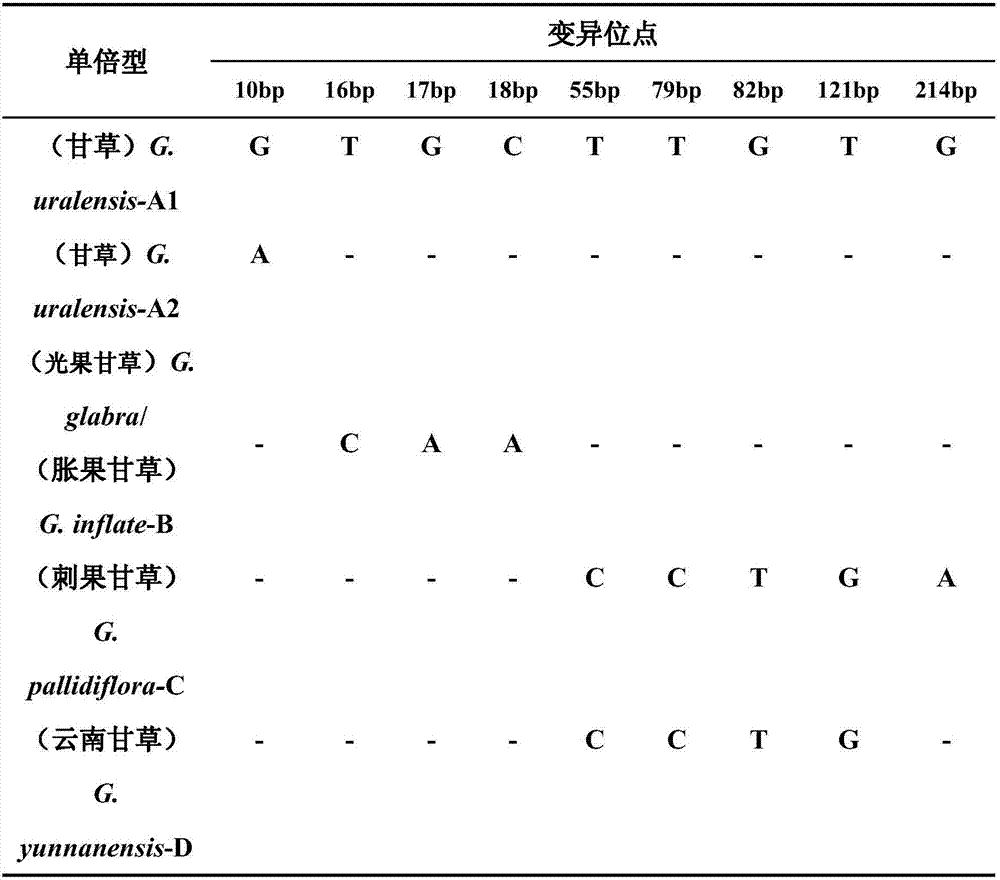
Medicinal liquorice root consanguinity identification method based on ITS2 sequence
InactiveCN107227367AEasy to distinguishEnsure safetyMicrobiological testing/measurementLiquoricesGLYCYRRHIZA EXTRACT
The invention discloses a medicinal liquorice root consanguinity identification method based on an ITS2 sequence. The method includes the identification on the consanguinity of medicinal liquorice roots and sibling species samples. The glycyrrhiza liquorice root species can be identified, so that various liquorice root species can be easily distinguished; the liquorice root medicinal condition disorder condition si avoided; the support is provided for ensuring the effectiveness and safety of liquorice root clinic medicine use.
Owner:GUIYANG COLLEGE OF TRADITIONAL CHINESE MEDICINE
PCR detection method for quickly detecting large coptotermes formosunus
InactiveCN101665828AProtect production securityQuick checkMicrobiological testing/measurementFluorescence/phosphorescenceForest industryFluorescence
The invention discloses a PCR detection method for quickly detecting large coptotermes formosunus, which comprises conventional PCR detection and real-time fluorescence PCR detection. The PCR detection method comprises the following steps: preparing a DNA template for a sample to be detected, designing a specific primer and / or a probe, establishing a PCR reaction system for PCR amplification, andobserving, recording and photographing on a gel imaging system. With the advantage of high speed, stability, reliability and sensitivity, the PCR detection method for quickly detecting large coptotermes formosunus is applied to the detection of different-grade samples of large coptotermes formosunus, and lays a foundation for China's coptotermes formosunus class in molecular classification and therapid identification of sibling species. The result of study can be directly applied to the quarantine identification of large coptotermes formosunus in inward and outward plant quarantine, and has important significance to the production safety protection of agriculture and forestry, biological balance maintenance, public health safety insurance in China.
Owner:中华人民共和国珠海出入境检验检疫局
DNA bar code standard gene used for identifying culex tritaeniorhynchus and applications
InactiveCN105420386AImprove accuracyPromote quality improvementMicrobiological testing/measurementDNA/RNA fragmentationDNA barcodingCulex tritaeniorhynchus
The invention belongs to the biotechnology field, and especially relates to a DNA bar code standard gene used for identifying culex tritaeniorhynchus and applications. The nucleotide sequence of the DNA bar code standard gene used for identifying culex tritaeniorhynchus is shown in the ID No.1 in a sequence table. The provided DNA bar code standard gene can be applied to identification whether a specimen to be detected is culex tritaeniorhynchus. The DNA bar code standard gene is suitable for practical needs of border port health quarantine work, and a relatively stable and accurate molecule identification method is established by utilization of the DNA bar code technology and aimed to culex tritaeniorhynchus. Defected samples can be identified, culex tritaeniorhynchus and sibling species can be distinguished and identified well, and the non-imago phase of culex tritaeniorhynchus can be identified accurately. The provided DNA bar code standard gene has important promoting effects on raising accuracy of culex tritaeniorhynchus classification identification and promoting health supervision work quality improvement.
Owner:宋锋林 +1
Molecular specificity labeling primer and method for identifying European boletus sibling species
InactiveCN103114149ARapid molecular identificationSimple methodMicrobiological testing/measurementDNA/RNA fragmentationForward primerMolecular identification
The invention provides a molecular specificity labeling primer capable of identifying European boletus sibling species B.appendiculatus and B.subappendiculatus, and a method for quickly identifying the European boletus sibling species B.appendiculatus and B.subappendiculatus by using the primer. The nucleotide sequence of the molecular specificity labeling primer is represented as follows: a forward primer: 5'-AGAAAAGCTCAGCCCGCTCAAA-3', and a reverse primer: 5'-ACTGTCGCTGGCCCCTCCCACTCTG-3'. According to the invention, the beneficial effects are as follows: the molecular specificity labeling primer is capable of quickly performing molecular identification on the European boletus sibling species B.appendiculatus and B.subappendiculatus; and the method is simple, fast and precise, and is an effective assistance for distinguishing two species of the boletus according to morphological characteristics.
Owner:ZHEJIANG FORESTRY ACAD
Molecular specificity marker primer and method for distinguishing Chinese bolete and European bolete
InactiveCN103173541AStrong subjectivityAccurate methodMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationBiotechnology
The invention provides a molecular specificity marker primer for distinguishing Chinese bolete B. roseoflavus and European bolete B. appendiculatus, and a method for quickly distinguishing Chinese bolete B. roseoflavus and European bolete B. appendiculatus by utilizing the primer. The nucleotide sequence of the molecular specificity marker primer is as follows: the upstream primer is 5'-CTTGCTCAGCCCAACTCAACG-3'; and the downstream primer is 5'-ATCTCTGGGTGGAGCATGTGCACG-3'. The molecular specificity marker primer and the method have the beneficial effects that quick molecular identification can be carried out on sibling species Chinese bolete B. roseoflavus and European bolete B. appendiculatus through the molecular specificity marker primer, and the method is simple, quick and accurate and is an effective assistant for distinguishing two types of bolete through morphology.
Owner:ZHEJIANG FORESTRY ACAD
Method for comparative analysis of ancient chromosome evolution of sisoridae fishes
PendingCN111564180AFacilitate understanding the laws of evolutionExtensive Application ResourcesBiostatisticsSequence analysisGeneticsFishery
The invention relates to a comparative analysis method for ancient chromosome evolution of sisoridae fishes. According to the invention, a comparative genomic technology is utilized to analyze the ancient chromosomes of the sisoridae fishes, and evolution research on the chromosomes of the sisoridae fishes is beneficial for exploring the evolution rule of the genomes of the sisoridae fishes and provides basic data for the evolution research and resource protection of the sisoridae fishes. The method provided by the invention comprises the steps of construction of the chromosome sequence of glyptosternon maculatum, phylogenetic analysis, comparison with the chromosomes of sibling species, ancient chromosome construction and the like. The method is suitable for the sisoridae fishes in Tibetan, and can be widely applied to resource investigation and protection research of the sisoridae fishes in the Qinghai-Tibet Plateau.
Owner:西藏自治区农牧科学院水产科学研究所
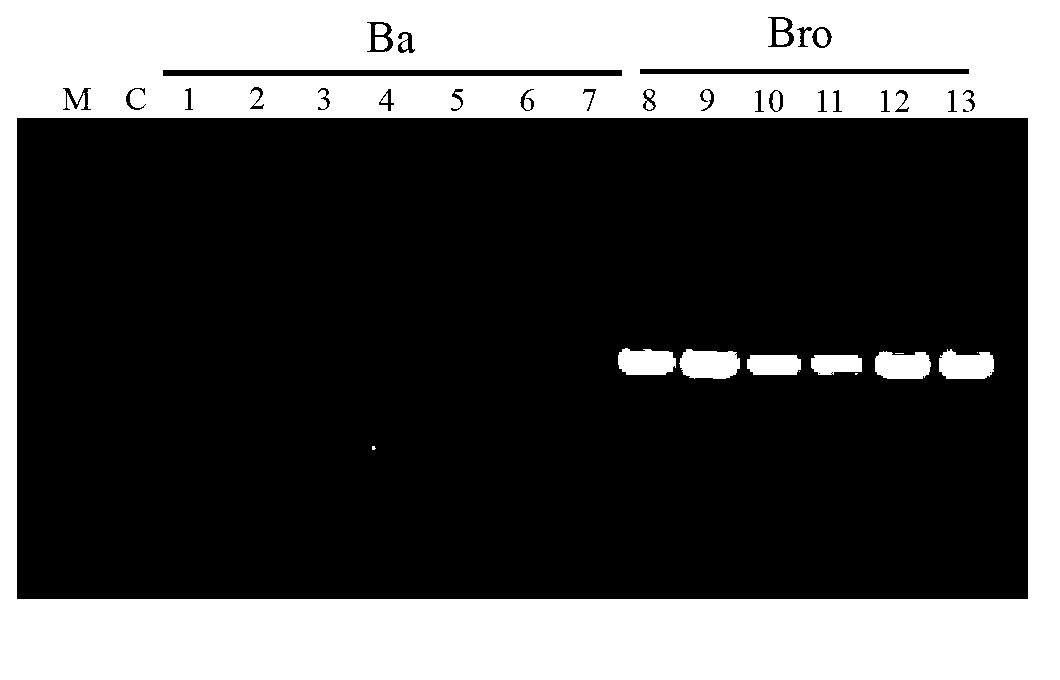
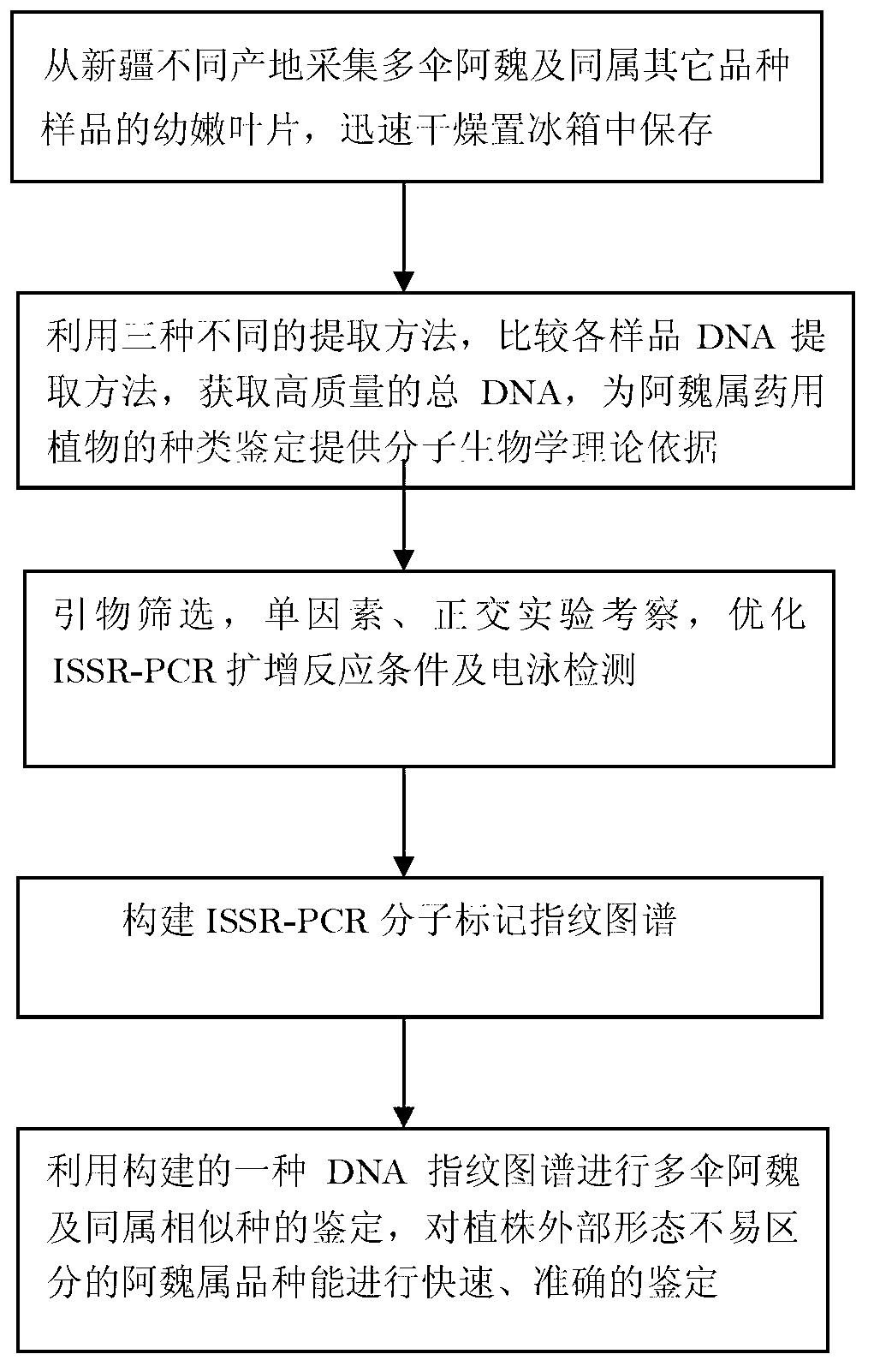
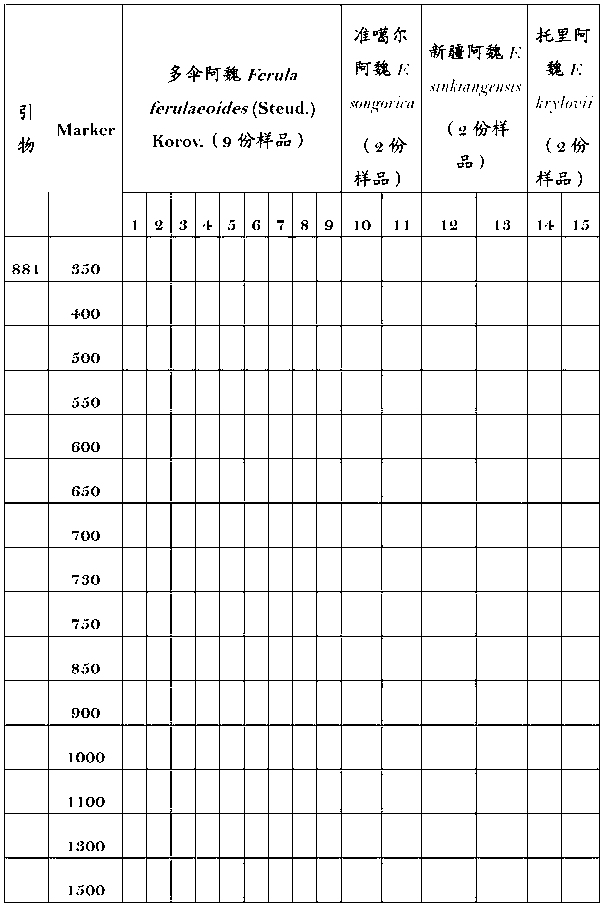
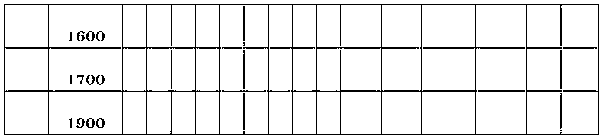
Molecular fingerprint identification method of ferula ferulaeoides and congeneric sibling species
InactiveCN103266171ARapid identificationAccurate identificationMicrobiological testing/measurementFerula ferulaeoidesBiology
The invention discloses a molecular fingerprint identification method of ferula ferulaeoides and congeneric sibling species, comprising the steps of creating a ferula ferulaeoides ISSR molecular marker optimization system, creating an ISSR (Inter Simple Sequence Repeat) fingerprint and identifying. The molecular fingerprint identification method specifically comprises the following steps: 1. collecting ferula ferulaeoides and other congeneric variety samples; 2. comparing genome DNA extraction methods of the samples; 3. optimizing ISSR-PCR (polymerase chain reaction) amplified reaction conditions and performing electrophoresis detection; 4. creating an ISSR molecular marker fingerprint; and 5. verifying the ferula ferulaeoides and the congeneric sibling species through the created DNA fingerprint. The molecular fingerprint identification method disclosed by the invention is beneficial to quickly and accurately identify ferula ferulaeoides varieties with external plant forms difficult to distinguish, saves time and cost and has important significance for accurately identifying medicinal material sources of ferula ferulaeoides.
Owner:XINJIANG MEDICAL UNIV
Method for quickly identifying pseudo-ginseng
InactiveCN102888456BRapid identificationAccurate identificationMicrobiological testing/measurement3-deoxyriboseAdulterant
The invention discloses a method for quickly identifying pseudo-ginseng. The method comprises steps as follows: 1) getting DNA (Deoxyribose Nucleic Acid) of a sample to be detected as a template, and amplifying the fragments containing the sequences shown as SEQ ID No.1 (Sequence Identification Number); and 2) sequencing the amplified products, analyzing the sequences as shown in SEQ ID No.1; if the bit 140 beginning from 5' end in the sequences as shown in SEQ ID No.1 is T or the bit 207 is T, identifying that the sample is pseudo-ginseng; and if the bit 140 and the bit 207 beginning from the 5' end in the sequence as shown in SEQ ID No.1 are not T, identifying that the sample is not pseudo-ginseng. With the adoption of the method disclosed by the invention, the raw pseudo-ginseng, the sibling species of the raw pseudo-ginseng, the crude medicine, the raw medicinal slices, the powder and the adulterant can be quickly and accurately identified.
Owner:INST OF MEDICINAL PLANT DEV CHINESE ACADEMY OF MEDICAL SCI
PCR (Polymerase Chain Reaction) primer and identification method of athetis lepigone and sibling species thereof
InactiveCN103266174ASolve the two-point committee mothSolving the puzzle of its morphologically similar speciesMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionGenomic DNA
The invention relates to a PCR (Polymerase Chain Reaction) primer and an identification method of an athetis lepigone and a sibling species thereof. The identification method comprises the following steps of: (1) extracting the genomic DNA of the athetis lepigone and the sibling species thereof; (2) carrying out PCR amplification on the mitochondrion COI (Cytochrome Oxidase I) gene of the genomic DNA by taking the genomic DNA of the athetis lepigone and the sibling species thereof as a template; (3) carrying out enzyme digestion on a PCR product prepared from the step (2) by using a restriction enzyme RsaI to obtain an enzyme-digested product; and (4) carrying out agarose gel electrophoresis analysis on the enzyme-digested product prepared from the step (3). According to the invention, the restriction enzyme is a common restriction enzyme, a simple, convenient and stable enzyme digestion marker is provided for screening the athetis lepigone and the sibling species thereof, and an identification technology of the athetis lepigone and the sibling species thereof is explored and established, so that the foundation is established for the future identification, population dynamic monitoring and integrated prevention and control of the athetis lepigone and the sibling species thereof.
Owner:QINGDAO AGRI UNIV
PCR detection method for quickly detecting large coptotermes formosunus
InactiveCN101665828BProtect production securityQuick checkMicrobiological testing/measurementFluorescence/phosphorescenceForest industryFluorescence
The invention discloses a PCR detection method for quickly detecting large coptotermes formosunus, which comprises conventional PCR detection and real-time fluorescence PCR detection. The PCR detection method comprises the following steps: preparing a DNA template for a sample to be detected, designing a specific primer and / or a probe, establishing a PCR reaction system for PCR amplification, andobserving, recording and photographing on a gel imaging system. With the advantage of high speed, stability, reliability and sensitivity, the PCR detection method for quickly detecting large coptotermes formosunus is applied to the detection of different-grade samples of large coptotermes formosunus, and lays a foundation for China's coptotermes formosunus class in molecular classification and the rapid identification of sibling species. The result of study can be directly applied to the quarantine identification of large coptotermes formosunus in inward and outward plant quarantine, and has important significance to the production safety protection of agriculture and forestry, biological balance maintenance, public health safety insurance in China.
Owner:中华人民共和国珠海出入境检验检疫局
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com