Patents
Literature
135 results about "Duplex pcr" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Primers, probes and detection kits for detection of porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus
InactiveCN102277454AShorten the timeSave costsMicrobiological testing/measurementFluorescence/phosphorescenceDuplex pcrDiarrhea
The invention discloses a primer, a probe and a detection kit for detecting porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. The detection primers and probes are SEQ ID NO: 1 to SEQ ID NO: 6 in the sequence table, wherein the sequences SEQ ID NO: 1 and SEQ ID NO: 2 are sense primers and antisense primers for detecting porcine transmissible gastroenteritis virus respectively, and the sequence SEQ ID NO: 3 For detecting the fluorescent probe of porcine transmissible gastroenteritis virus, sequence SEQIDNO: 4 and SEQIDNO: 5 are sense primer and antisense primer for detecting porcine epidemic diarrhea virus respectively, and sequence SEQIDNO: 6 is the detection primer of porcine epidemic diarrhea virus fluorescent probe. The invention also provides detection kits for porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. The primers and probes selected by the present invention have very strong specificity. The total viral RNA extracted from porcine diarrhea does not need to be transcribed into cDNA first. The synthesis of the first strand of cDNA and double PCR are completed in one step, and two viruses can be detected at one time. ,Improve efficiency.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Dual PCR detection probe and kit for duck adenovirus 2 and duck adenovirus A
InactiveCN107058634AEquivalent sensitivitySimplify operating proceduresMicrobiological testing/measurementDNA/RNA fragmentationDuplex pcrMicrobiology
The invention provides a dual PCR detection probe and kit for duck adenovirus 2 and duck adenovirus A, and belongs to the field of epizootiology. Two pairs of primers which are shown in SEQ ID NO.1-4 are adopted, a dual PCR detection method through which differential diagnosis can be conducted on duck adenovirus 2 and duck adenovirus A in a duck flock is built, a basis is laid for developing an epidemiological survey of adenovirus infection types and scientific prevention and control of relevant diseases in the duck flock, and thus the dual PCR detection probe and kit for duck adenovirus 2 and duck adenovirus A have very important study significance.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
Method for identifying fast and slow feather molecules of chickens
InactiveCN101619354AAccurate identificationShorten the establishment timeMicrobiological testing/measurementZ chromosomeDuplex pcr
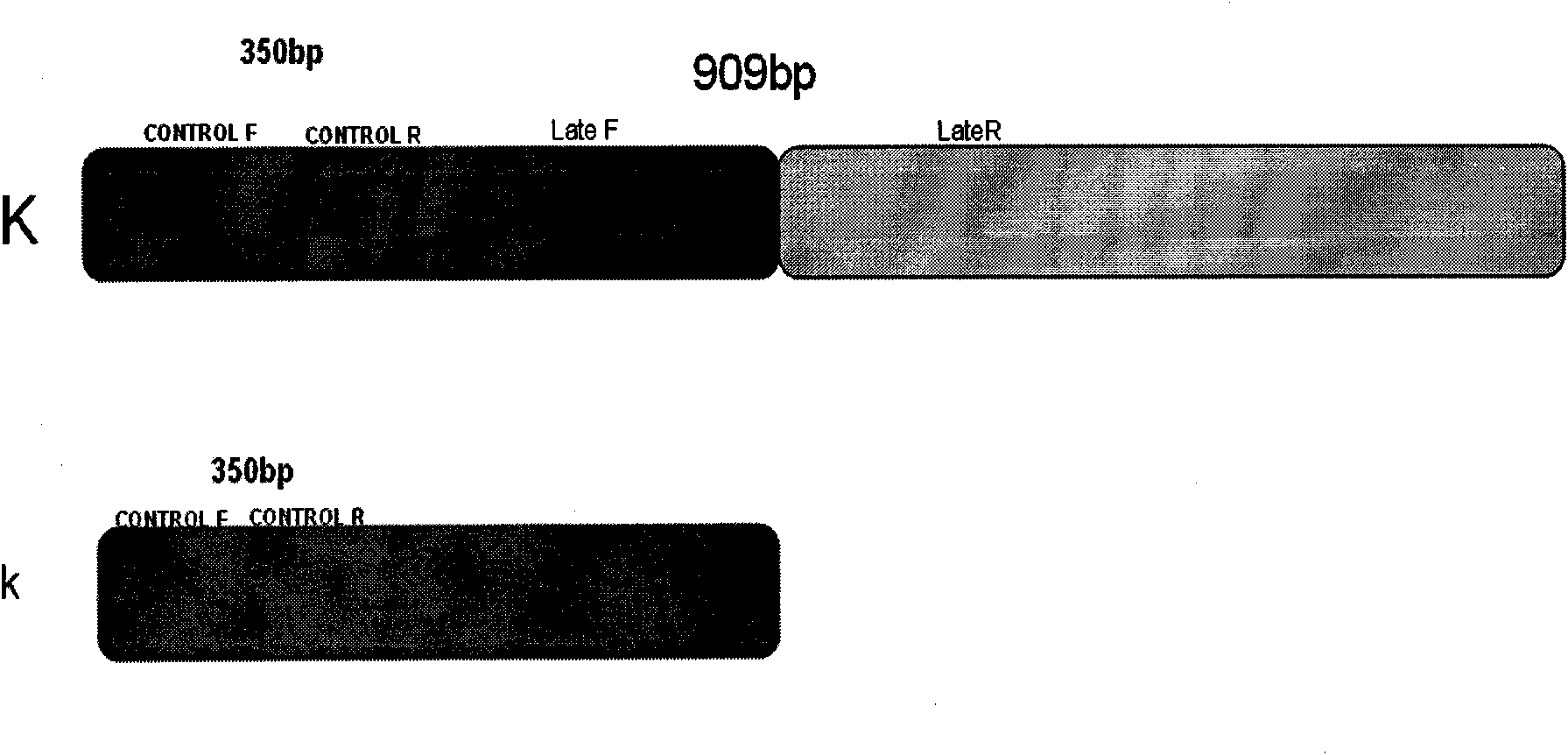
The invention belongs to the technical field of animal breeding, in particular to a method for identifying fast feather type and slow feather type of chickens by applying double PCR. The method comprises the following steps: (1) designing two pairs of primers by utilizing a correlation sequence of a K gene locus found on a Z chromosome of a chicken according to structures of alleles of the fast and slow feather type; (2) extracting DNAs of chicken blood samples of the known fast and slow feather type to carry out a primer specificity test, and identifying the chicken to be a fast feather type or a slow feather type according to the size and the existence of an amplified DNA segment. When only one 350bp band is amplified, the chicken is judged to be a fast feather type, and when two bands of 350bp and 909bp are amplified, the chicken is judged to be a slow feather type. The invention also discloses a reliable checking method of the identifying method and applies the checking method to a breeding process of a self identification male and female matched system of the fast and slow feathers. The invention can shorten the breeding time of the self identification male and female matched system of the fast and slow feathers and save the breeding cost.
Owner:HUAZHONG AGRI UNIV
Molecular detection method of Brassica napus self-incompatible S-locus haplotype
InactiveCN103451283AEasy to useSolve the problem of low coordinationMicrobiological testing/measurementDNA/RNA fragmentationBrassicaDuplex pcr
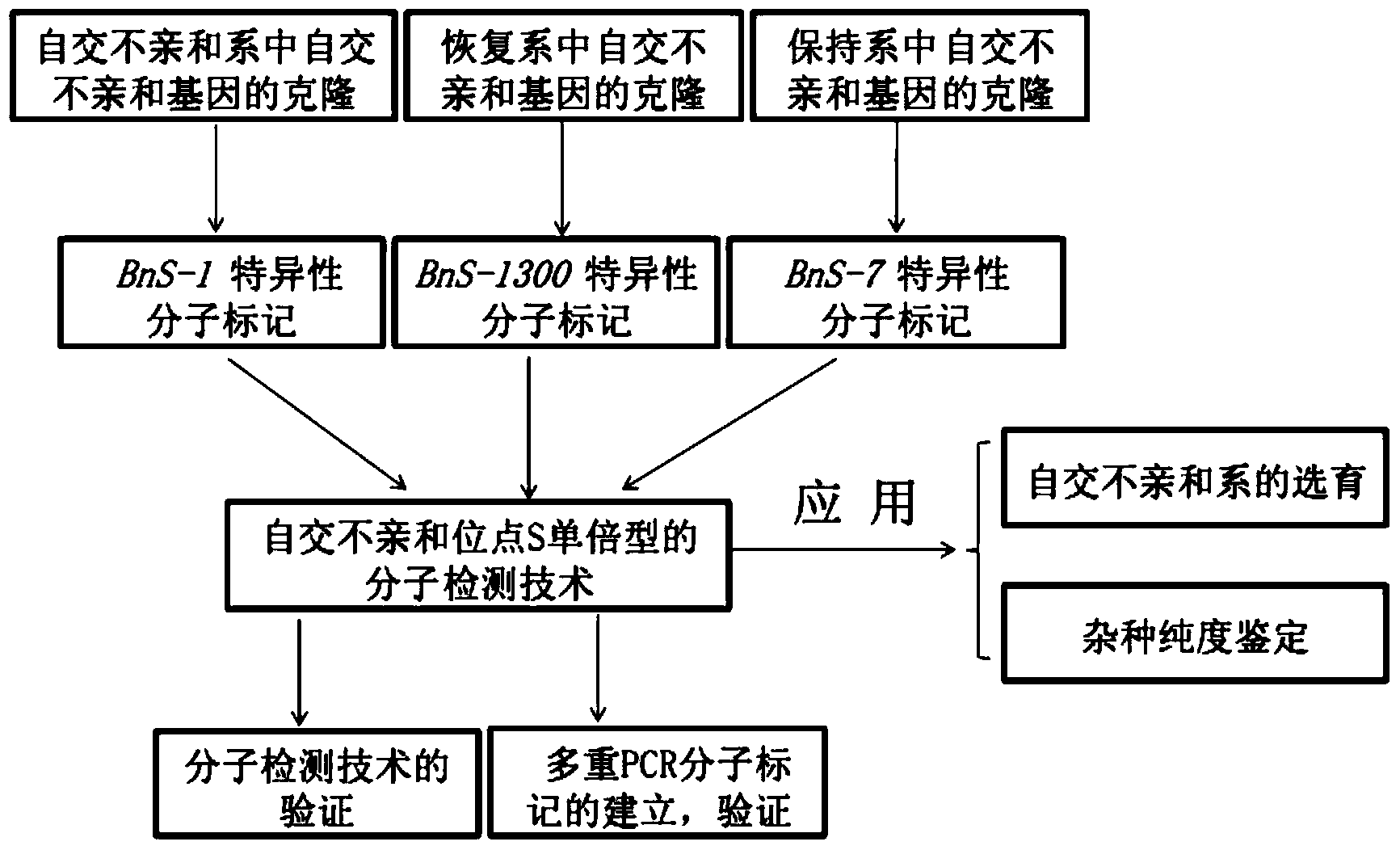
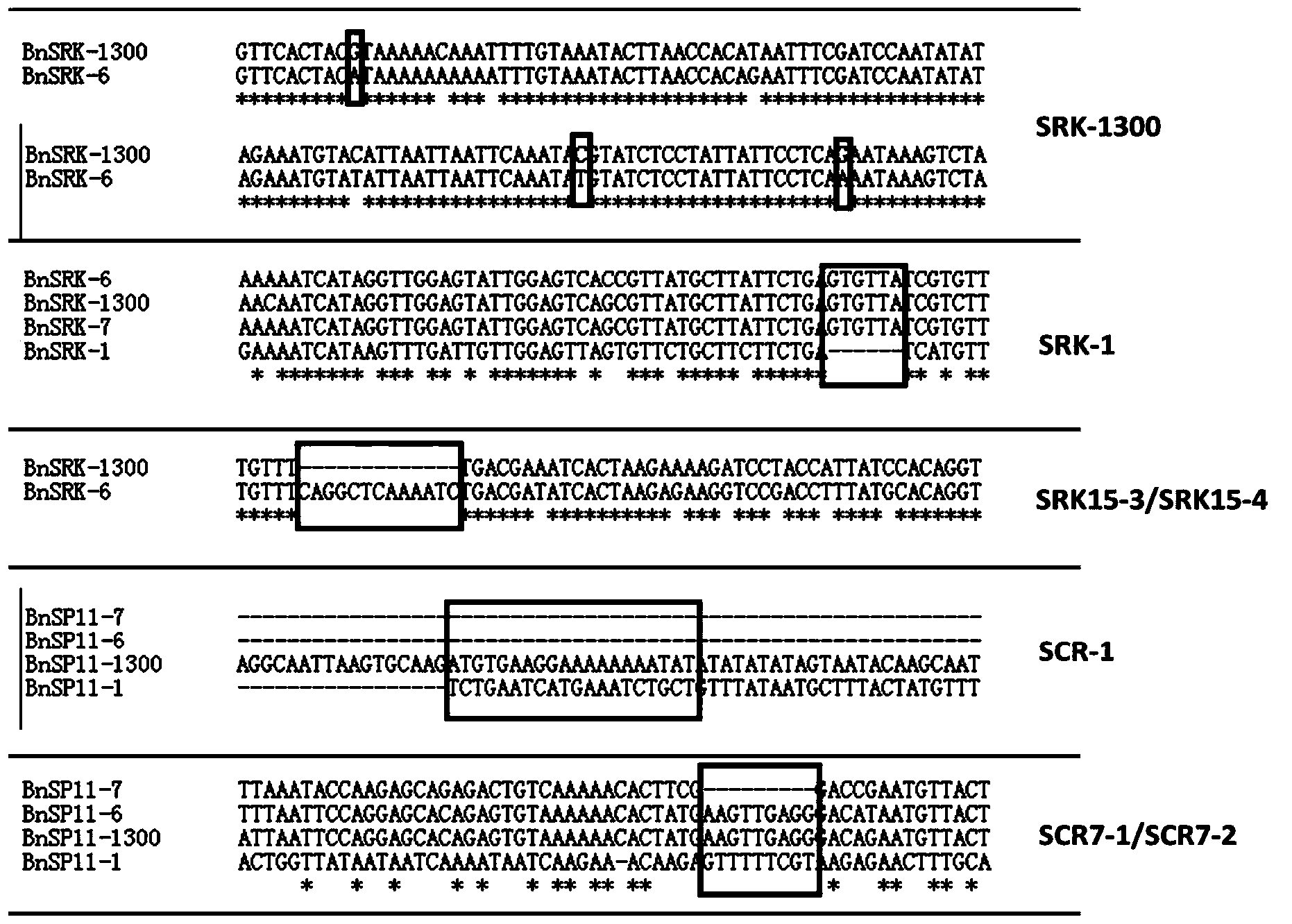
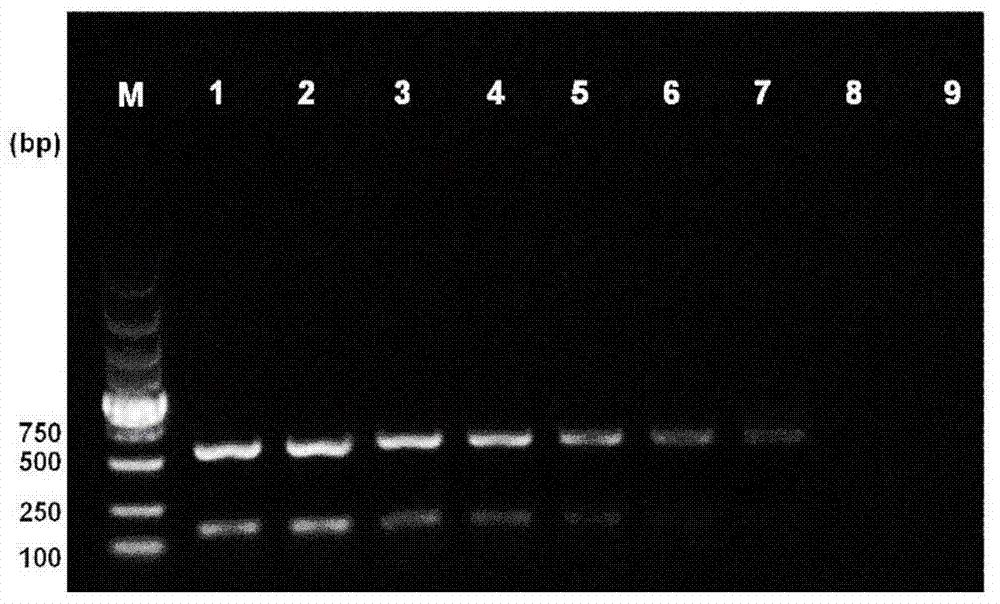
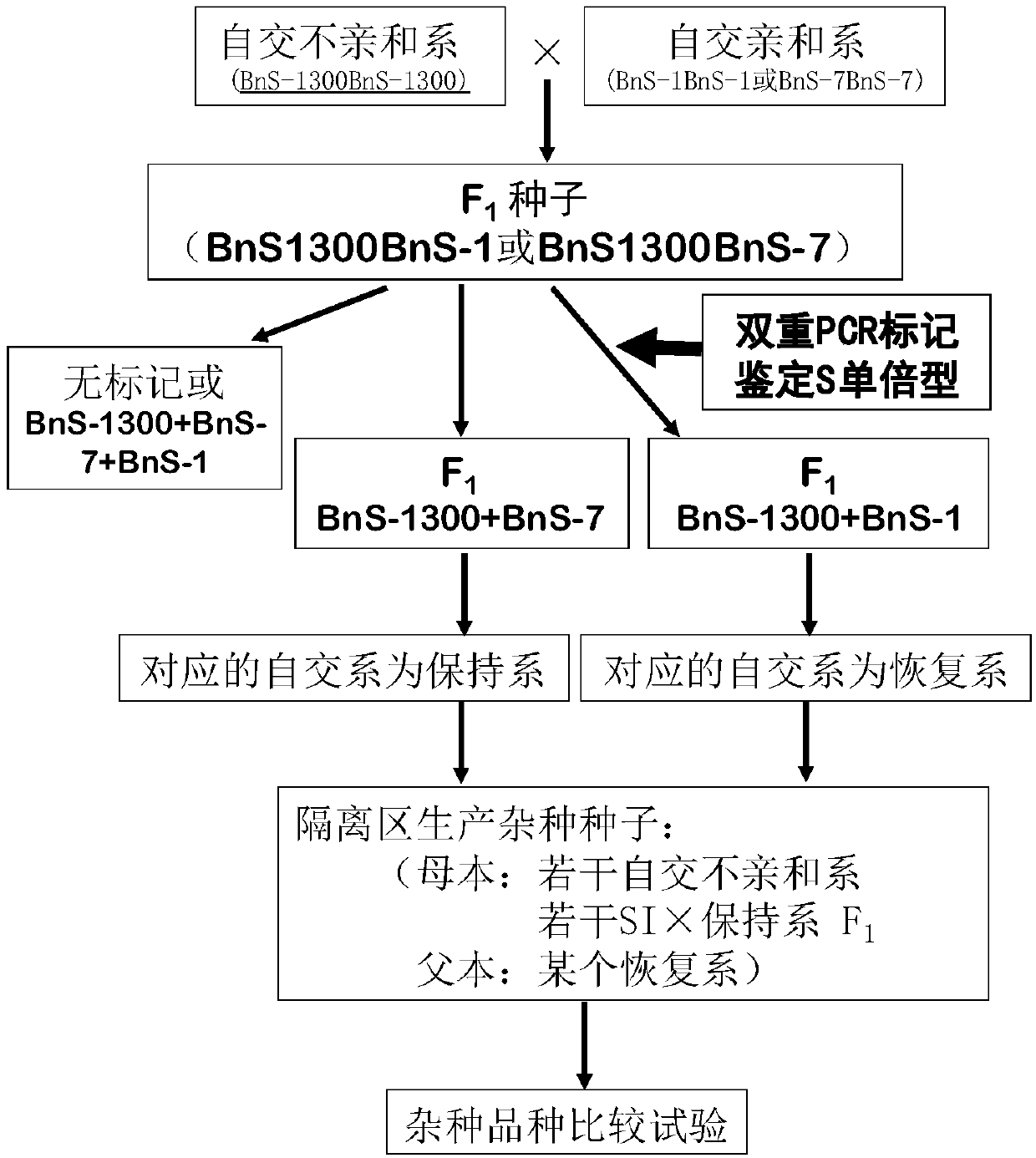
The invention belongs to the field of molecular breeding of rapeseeds, and relates to a molecular detection method of Brassica napus self-incompatible S-locus haplotype, a molecular marker and application. An SCAR (sequence characterized amplified regions) molecular marker system used for identifying the Brassica napus self-incompatible S-locus haplotype is prepared and includes four SCAR molecular markers, namely, SRK1-1, SCR1-1, SRK-1300 and SCR7-2 which can be combined to obtain two pairs of double PCR (polymerase chain reaction) markers as SRK1-1 and SRK-1300 as well as SCR7-2 and SRK-1300; the PCR marker combination can be used in a combination manner or independent manner; the DNA (deoxyribonucleic acid) sequences of the PCR marker combination are shown as SEQ ID NO: 6, SEQ ID NO:7, SEQ ID NO: 8 and SEQ ID NO: 9. According to the invention, a practical marker and an utilization method are provided for the molecular breeding of the rapeseeds.
Owner:HUAZHONG AGRI UNIV
Gene methylation detection method
InactiveCN104131070AImprove accuracyGood repeatabilityMicrobiological testing/measurementReference genesDuplex pcr
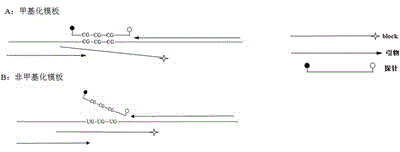
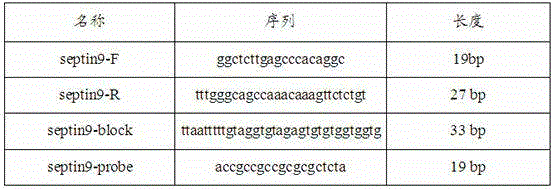
The invention discloses a gene methylation detection method. The method comprises the steps of: subjecting a to-be-detected DNA sample, a negative control, a positive control and a no template control to PCR amplification reaction, with the DNA sample in the PCR amplification reaction being bisulfite modified DNA, carrying out duplex PCR amplification reaction on a detection gene and a reference gene, and interpreting a parameter range to judge the methylation condition of the detection gene of the to-be-detected DNA sample. By the mode, the gene methylation detection method provided by the invention has the advantages of strong specificity, duplex reaction, high sensitivity, small pollution, simple and quick operation, and safety, etc., and the detection result has good accuracy and repeatability. Thus, the method has certain auxiliary diagnosis effect on early detection of colorectal cancer and other cancers.
Owner:JIANGSU MICRODIAG BIOMEDICINE TECH CO LTD
Duplex PCR authentication method of cordyceps sinensis original powder
ActiveCN103233062AAmplification conditions are easyPrimer specificityMicrobiological testing/measurementDuplex pcrHousekeeping gene
The invention discloses a duplex PCR authentication method of cordyceps sinensis original powder. The method adopts a technical scheme that cordyceps sinensis original powder genome DNA is adopted as a template; with PCR primers designed by the invention, a single-tube duplex PCR reaction is carried out, and characteristic housekeeping genes of a cordyceps sinensis strain and a host hepialus are simultaneously amplified, wherein actual amplified fragments are respectively 320bp and 136bp; and through agarose gel electrophoresis detection, product authenticity is determined. With the method provided by the invention, cordyceps sinensis original powder can be specifically identified, and sample authenticity detection can be completed simply with three steps of DNA extraction, PCR amplification, and electrophoresis detection. The operation is simple and is easy to command, and accuracy is high.
Owner:CHINA NAT RES INST OF FOOD & FERMENTATION IND CO LTD +1
Duplex polymerase chain reaction (PCR) method for rapidly identifying duck circovirus genotype
InactiveCN104059998AIncreased sensitivityAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationDuplex pcrGenotype
The invention provides a duplex polymerase chain reaction (PCR) method for rapidly identifying duck circovirus genotype. The method comprises the following steps: comparing sequences DuCV-1 and DuCV-2 released on reference GenBank, selectively and respectively designing a pair of general specific primers SEQ1 / SEQ2 aiming at DuCV, a type 1 duck circovirus specific primer SEQ3 and a type 2 duck circovirus specific primer SEQ4 in a conserved region of the DuCV-1 and DuCV-2. According to the method, novel specific primers are designed and screened, various parameters in the reaction process are optimized, the dual PCR sequence-based typing method aiming at the duck circovirus established by utilizing the primers is high in specificity and high in sensitivity, and the sequences DuCV-1 and DuCV-2 can be rapidly and accurately identified.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Dual PCR primer, detection method and kit for detecting porcine circovirus type 2 and circovirus type 3
InactiveCN107955839AEasy to detectImprove featuresMicrobiological testing/measurementMicroorganism based processesDuplex pcrCircovirus
The invention provides a dual PCR primer, detection method and kit for detecting porcine circovirus type 2 and circovirus type 3. The dual PCR primer is designed according to a highly-conserved specific sequence for detecting porcine circovirus type 2 and circovirus type 3, and the detection method for monotube synchronous detecting of porcine circovirus type 2 and circovirus type 3 is established. The kit has the advantages of quickness, simplicity, convenience and high specificity, sensitivity and reliability, sample analysis on a large scale can be performed at the same time, whether a sample is infected with porcine circovirus type 2 or the porcine circovirus type 3 or the two can be identified and diagnosed through single reaction, powerful technical support is provided for monitoring, preventing and controlling porcine circovirus epidemic, and the kit has a good application prospect.
Owner:SOUTH CHINA AGRI UNIV
Potato rot nematode rDNA-ITS sequence, specific detection primer and one-step dual PCR detection method
The present invention is potato rot stem nematode rDNA-ITS sequence, specific detection primer and one-step double PCR detection method. The potato rot stem nematode rDNA-ITS has the nucleotide sequence as shown in SEQ No. 1 or SEQ No. 2. The specific detection primer SSsj designed based on the rDNA-ITS sequence and general purpose primer AB28 are combined and added with internal standard D3A and D3B for one-step double PCR detection. The PCR amplification result of potato stem nematode can obtain specific 623 bp segment and 345 bp segment, but that of other stem nematode can obtain specific 345 bp segment only. The present invention provides fast molecular detection method for stem nematode detection and identification and thus provides theoretic reference for comprehensive control of stem nematode disease.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
Identification method for determining anastomosis groups of rhizoctonia solani
InactiveCN101792795ARapid and Accurate Molecular Detection MethodsMicrobiological testing/measurementBiotechnologyDuplex pcr
The invention relates to an identification method for determining anastomosis groups of rhizoctonia solani, which belongs to field of biotechnology. The identification method comprises the steps that: on the basis of processes of the extraction on total DNA of bacterial strains in eight different anastomosis groups of rhizoctonia solani, PCR amplification, the measurement and analysis of rDNA-ITS sequences and the like, basic group loci for identifying the bacterial strains in the different anastomosis groups of the rhizoctonia solani are found, high-specificity identification primers for identifying the bacterial strains in the different anastomosis groups are respectively designed, and the high-specificity identification primers are combined with a general primer ITS4 and a general primer ITS1 for detection by a one-step double PCR method, so that DNA fragments with specificity of 500 bp and 700 bp are obtained, and the identification of the high-specificity PCR of the bacterial strains in the different anastomosis groups is performed successfully. The identification method provides a quick and accurate molecular detection method for the identification of the rhizoctonia solani and provides theoretical reference for making the comprehensive treatment decision of multiple diseases such as banded sclerotial blight, rhizoctonosis and the like caused by formulating the rhizoctonia solani.
Owner:北京达成生物科技有限公司
Duplex PCR (polymerase chain reaction) detection kit for duck Newcastle disease virus and duck circovirus
ActiveCN102618668ASimple and fast operationIncreased sensitivityMicrobiological testing/measurementDNA/RNA fragmentationDuplex pcrNewcastle disease virus NDV
The invention discloses a duplex PCR (polymerase chain reaction) detection kit for duck Newcastle disease virus and duck circovirus, and particularly relates to a primer group for detecting Newcastle disease virus and duck circovirus, which comprises a first primer, a second primer, a third primer and a fourth primer. Nucleotide sequences of the first primer, the second primer, the third primer and the fourth primer are respectively a first sequence, a second sequence, a third sequence and a fourth sequence in a sequence list. Testing shows that the two pairs of primers are designed to establish a duplex PCR detection method for Newcastle disease virus and duck circovirus, and the duplex PCR technique capable of detecting two pathogens, namely the Newcastle disease virus and the duck circovirus has the advantages of simplicity in operation, high sensitivity, high specificity, high repeatability and the like.
Owner:GUANGXI VETERINARY RES INST
Duplex PCR (polymerase chain reaction) detection primer and kit for quickly distinguishing porcine circoviruses type 2 and type 3
InactiveCN108531656AEasy to operateQuick checkMicrobiological testing/measurementMicroorganism based processesDuplex pcrSterile water
The invention belongs to the technical field of animal virology and molecular biology and discloses a duplex PCR (polymerase chain reaction) detection primer and kit for quickly distinguishing porcinecircoviruses type 2 and type 3. The kit comprises primer sequences shown as SEQ ID1-4, 2*F8 Fastlong PCR MasterMix, a cDNA template and sterile water. The duplex PCR detection kit for quickly distinguishing the porcine circoviruses type 2 and type 3 has advantages that by duplex PCR amplification, two types of DNA viruses similar in lesion can be detected in one time, the total detection time iscontrolled to be about two hours, a detection method is easy in operation, convenient and quick, and quick detection can be realized.
Owner:GUANGXI VETERINARY RES INST
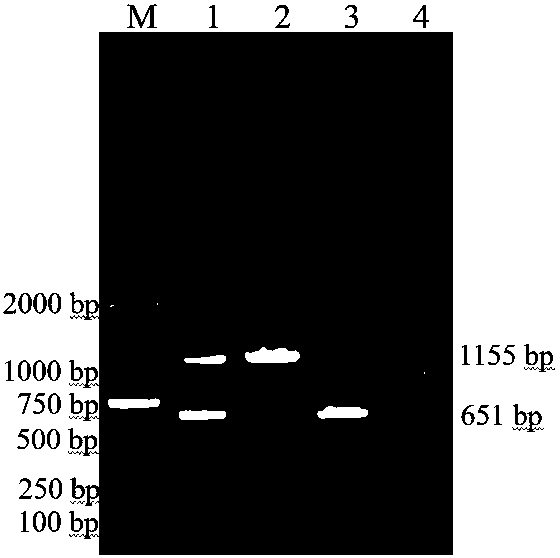
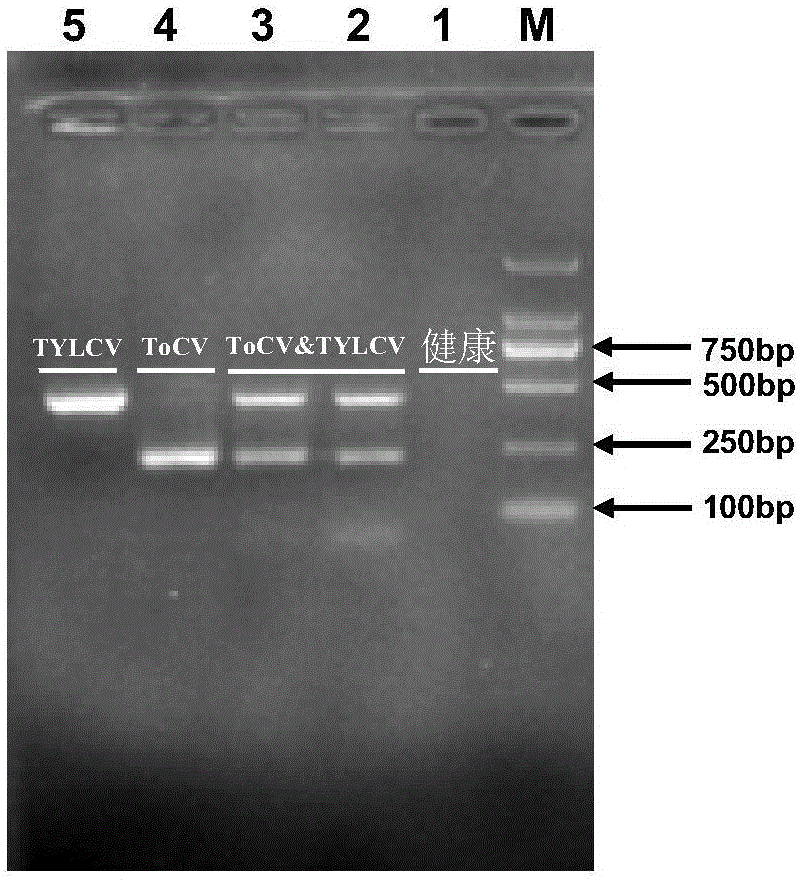
Duplex PCR (polymerase chain reaction) primer and method for identifying tomato chlorosis viruses and tomato yellow leaf curl viruses
InactiveCN105087567AImprove detection efficiencyReduce dosageMicrobiological testing/measurementDNA/RNA fragmentationDuplex pcrAgricultural science
The invention relates to a duplex PCR (polymerase chain reaction) primer and a method for identifying tomato chlorosis viruses and tomato yellow leaf curl viruses. The method comprises the following steps that (1) plant total RNA (ribose nucleic acid) is extracted; (2) the plant total RNA is used as a template for synthesizing cDNA (complementary deoxyribonucleic acid) through reverse transcription; (3) the cDNA is used as a template for performing PCR amplification on a nucleotide sequence coded by ToCV and TYLCV capsid protein; (4) a PCR product prepared in the step (3) is subjected to agarose gel electrophoresis analysis. The invention relates to a PCR primer combination. A duplex PCR method is used for performing ToCV and TYLCV identification; the result is accurate; the method is fast, simple and convenient; the detection cost is reduced; the virus detection efficiency is greatly improved; good application prospects are realized.
Owner:QINGDAO AGRI UNIV
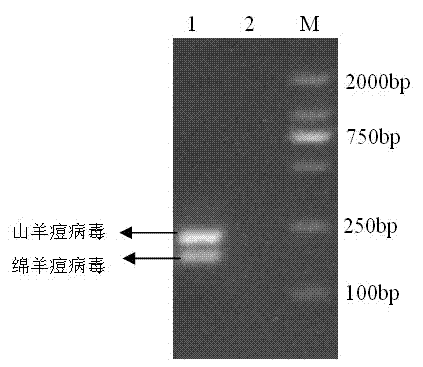
Goat pox virus and sheep pox virus dual-PCR (Polymerase Chain Reaction) detection kit and detection method
ActiveCN102367492AEasy to identifyPromote amplificationMicrobiological testing/measurementMicroorganism based processesDuplex pcrPcr method
The invention discloses a goat pox virus and sheep pox virus dual-PCR (Polymerase Chain Reaction) detection kit and a dual-PCR method for rapidly identifying and detecting goat pox virus and sheep pox virus. One pair of PCR primers is respectively designed according to genome sequences of the goat pox virus and the sheep pox virus, and corresponding specific fragments can be obtained through amplification, thus detection and identification of the goat pox virus and the sheep pox virus are realized. By using the dual-PCR technology, the goat pox virus and the sheep pox virus can be simultaneously detected by using a single tube, thus detection cost and workload of goat pox and sheep pox are reduced, and identification detection of the goat pox and the sheep pox is realized. According to the invention, a dual-PCR method with the advantages of strong specificity, high sensitivity, time and labor saving and easiness in observing results is established, and the advantages of convenience for operation, strong specificity, high sensitivity, high repeatability and no complex post-treatment are achieved.
Owner:重庆海关技术中心 +1
Primer pair for detecting citrus canker pathogenic bacteria and detection method thereof
InactiveCN101712983AImprove accuracyEasy to judgeMicrobiological testing/measurementMicroorganism based processesDuplex pcrPathogenic bacteria
The invention relates to the biological detection technology, in particular to primers for detecting citrus canker pathogenic bacteria and a detection method thereof. The invention aims to provide the primers and method for dual PCR-DHPLC detection of the citrus canker pathogenic bacteria. The primers for detecting the citrus canker pathogenic bacteria of the invention have good primer specificity and high detection sensitivity; and the detection method of the invention has high accuracy and high sensitivity and can quickly and simply judge whether a sample has the citrus canker pathogenic bacteria or not, thereby ensuring the safety in import and export.
Owner:中华人民共和国湖南出入境检验检疫局
Primer composition for detecting legionella and kit using the same
ActiveCN104131089AStrong specificityIncreased sensitivityMicrobiological testing/measurementMicroorganism based processesDuplex pcrAlveolar lavage fluid
The invention provides a primer composition for detecting legionella in clinical and environmental specimens. The primer composition comprises a pair of primers (shown in the Seq ID No.1-2) for PCR specific amplification of a gene mip and a pair of primers (shown in the Seq ID No.3-4) for PCR specific amplification of 16S rRNA. The invention also provides a detection kit containing the primer composition. A test on 320 groups of phlegm and alveolar irrigation solutions of pneumonia patients and 210 groups of environmental water specimens proves that the primer composition has singularity of 100% and a lowest detection limit of 2*10<1> cfu / ml. The dual PCR system can be successfully used in bacterial strain identification and clinical and environmental specimen detection. The PCR detection method built based on the dual PCR system has the characteristics of simpleness, fastness and reliability and can be widely used in epidemiology preliminary investigation, environmental water monitoring and clinical specimen detection.
Owner:ICDC CHINA CDC
Double PCR (polymerase chain reaction) rapid detection method for escherichia coli O157:H7 and kit
InactiveCN102154488AStrong specificityAvoid false positivesMicrobiological testing/measurementMicroorganism based processesEscherichia coliDuplex pcr
The invention discloses a double PCR (polymerase chain reaction)) rapid detection method for escherichia coli O157:H7 and a kit; the method comprises the following steps of: selecting a 0517 specific antigen gene rfbE and a coding H7 flagellum gene Flic of escherichia coli as amplified target genes, and simultaneously taking homology of different serotype nucleotides into consideration, and designing two pairs of specific primers which are used for rapidly detecting escherichia coli O157:H7 by PCR (polymerase chain reaction). The method is simple and convenient in operation, saves time, has high specificity and good sensitivity, only can detect the escherichia coli O157:H7 and is not affected by non-F7 serotype and other related bacterium of the escherichia coli. The kit which is researched based on the double PCR (polymerase chain reaction)) rapid detection method is suitable for bulk detection of various samples and provides a new technical means for rapidly detecting the escherichia coli O157:H7.
Owner:GUANGXI VETERINARY RES INST
Primer for detecting separation purity of X, Y spermatozoon of cattle
InactiveCN101195842ALow costEasy to operateSugar derivativesMicrobiological testing/measurementDuplex pcrStatistical analysis
The invention provides a primer for detecting the X and Y sperm separation purity. Aiming at a sex-determining gene Sry on a bull Y chromosome, the primer is designed through the PCR mismatching technology. The fragment size can be amplified by 295 bp through the primer, in order to prevent false positive from appearing, a pair of internal control primers C34 is designed through the invention according to a bull autosome 3 reported sequence, the fragment size is amplified by 208bp, dual PCR amplification is performed to single bull sperm through the two pairs of primers, and then the final evaluation is performed to the sperm separation purity according to the statistical analysis to the detection result. The invention provides the technology used for identifying the bull X and Y sperm separation purity with low cost and simple, rapid, and accurate operation, and provides reliable technical support for the popularization and the application of the subsequent sexing semen and the optimization of a sperm separation method.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
Siniperca chuatsi ranairidovirus and rhabdovirus duplex PCR detection kit and detection method
PendingCN111254225AMultiple treatment timesQuick checkMicrobiological testing/measurementMicroorganism based processesDuplex pcrRhabdovirus carpio
The invention relates to a siniperca chuatsi ranairidovirus and rhabdovirus duplex PCR detection kit and detection method, and belongs to the technical field of PCR. The siniperca chuatsi ranairidovirus and rhabdovirus duplex PCR detection kit provided by the invention comprises primers, EasyTaq PCR SuperMix, a negative control solution and a positive control solution, wherein the primers comprisea primer designed by aiming at an MCP gene conserved region of the siniperca chuatsi ranairidovirus, and a primer designed by aiming at an N gene conserved region of the siniperca chuatsi rhabdovirus. The siniperca chuatsi ranairidovirus and rhabdovirus duplex PCR detection kit and detection method provided by the invention can be used for fast, efficiently and accurately detecting the sinipercachuatsi ranairidovirus and the siniperca chuatsi rhabdovirus at the same time; the time and the labor are saved; more treatment time is won for sick siniperca chuatsi; and important significance is realized on subsequent study, prevention and control.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
Double detection method of porcine reproductive and respiratory syndrome virus and bovine viral diarrhea virus from pig
InactiveCN101935716AHigh utility valueIncreased sensitivityMicrobiological testing/measurementDuplex pcrBovine Viral Diarrhea Viruses
The invention relates to a double RT-PCR detection method of porcine reproductive and respiratory syndrome virus (PRRSV) and bovine viral diarrhea virus from a pig (BVDV), belonging to the biotechnology field. A pair of gene specific primers are respectively designed aiming to the PRRSV and the BVDV, the double RT-PCR detection method of the BVDV and the PRRSV is established, and minimum amounts of cDNA detection of the PRRSV and the BVDV are respectively 3.8*10<-4>ng and 7*10<-4>ng. Samples of a lung, a spleen, a lymph gland and the like of 75 piglets of Jiangsu Province are detected through the method to have a result that 55 are PRRSV positive and 14 are BVDV positive, wherein 12 PRRSVs and 12 BVDVs are mixedly infected, the detection result coincidence rate of the PRRSV and a single RT-PCR and the detection result coincidence rate of the BVDV and the single RT-PCR are respectively 89.3 percent and 92 percent, thereby proving that the established double RT-PCR detection method can be used for detecting the BVDV and the PRRSV in a clinical sample. Compared with a single detection method, the double RT-PCR detection method can save the time and the cost of an experiment and enhance the detection efficiency.
Owner:NANJING AGRICULTURAL UNIVERSITY
Duplex PCR method for detecting porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus
InactiveCN103952496AHigh sensitivityStrong specificityMicrobiological testing/measurementBiotechnologyDuplex pcr
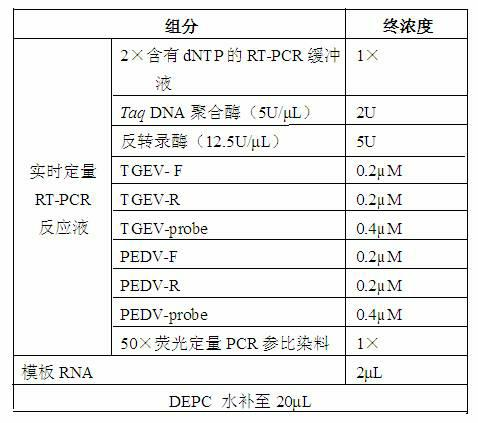
The invention belongs to the technical field of biology, and concretely relates to a duplex PCR method for detecting porcine transmissible gastroenteritis virus (TGEV) and porcine epidemic diarrhea virus (PEDV). The method comprises: designing two primers respectively amplifying the specific sequences of TGEV and PEDV, extracting RNA from positive intestinal canal tissue infected TGEV and PEDV, performing inverse transcription to obtain cDNA, and further taking cDNA as a template, so as to establish a multiplex PCR detection method capable of directly detecting two kinds of diarrhoea viruses from a sample once. The beneficial effects comprise that compared with presently reported PCR detection methods, the duplex PCR detection method is improved in detection efficiency and strong in specificity.
Owner:SHANGHAI JIAO TONG UNIV +1
Detection kit for aeromonas veronii and application method thereof
InactiveCN106399520AStrong specificityQuick checkMicrobiological testing/measurementMicroorganism based processesDuplex pcrSibling species
The invention provides a detection kit for aeromonas veronii and an application method thereof. In the technical scheme, gyrB gene is found to be a single-copy house-keeping gene through experiments and has more remarkable advantages than conventional 16S rRNA in the distinguishing and identifying of aeromonas veronii and sibling species thereof; and meanwhile, Aha gene is found to be stably exist in the aeromonas veronii and related to the toxicity thereof. Based on the beneficial discoveries, the gyrB gene and Aha gene are adopted as target genes to develop a duplex PCR detection method for aeromonas veronii; in the method, specific primers are designed for the two target genes, and a PCR reaction system and reaction conditions are determined. In the invention, the sensitivity and specificity of aeromonas veronii detection are high, the pelteobagrus fulvidraco-source pathogenic aeromonas veronii can be quickly and accurately detected, and the wrong detection and missed detection are effectively avoided. Meanwhile, repeated culture and redundant series of biochemical reactions are prevented, and the time, labor and cost are saved.
Owner:SOUTHWEST UNIVERSITY
Duplex PCR diagnostic kit for detection of contagious caprine pleuropneumonia pathogen and preparation and use methods thereof
InactiveCN102634601ADefinitive diagnosisrapidReduce complexityMicrobiological testing/measurementMicroorganism based processesDuplex pcrPositive control
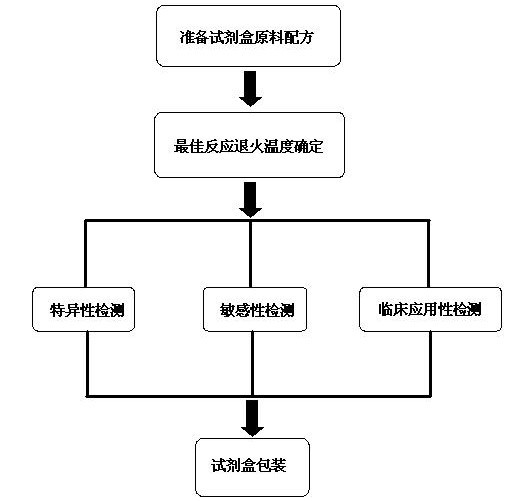
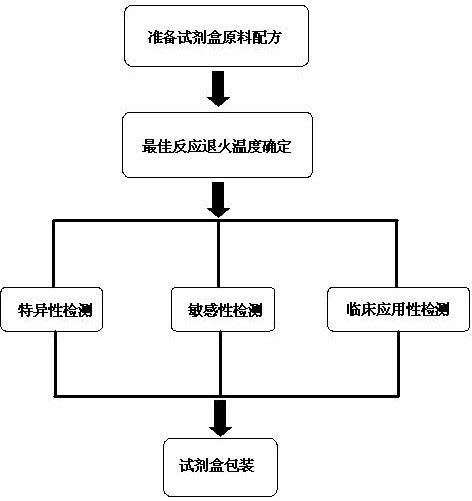
The invention discloses a raw material composition of a duplex PCR (polymerase chain reaction) diagnostic kit for detection of contagious caprine pleuropneumonia pathogen and a preparation method and a use method of the kit. The raw material composition comprises 1000-2000muL of PCR reaction liquid, 100-200muL of TaqDNA polymerase, 50-100muL of positive control, 50-100muL of negative control and 1-2mL of ultrapure water. The PCR reaction solution comprises a primer 1 mixture solution, a primer 2 mixture solution, a PCR reaction buffer and dNTPs (deoxyribonucleotide triphosphates), wherein the volume ratio of the four liquids in the PCR reaction solution is 2:2:5:1. The preparation method comprises the steps of determination of an optimum reaction annealing temperature, specific detection, sensibility test, clinical application detection and kit packaging. An effective method for rapid deterministic diagnosis of suspected goat pathogen and suspected cases of contagious caprine pleuropneumonia in goat farms is provided by the invention, providing technical support for the prevention and control of contagious caprine pleuropneumonia.
Owner:GUIZHOU UNIV
Duplex PCR detection kit and detection method for Listeria monocytogenes and Enterococcus faecium
ActiveCN104846067ADetection helpsAids in early diagnosisMicrobiological testing/measurementMicroorganism based processesLower limitDuplex pcr
The invention relates to a duplex PCR detection kit for Listeria monocytogenes and Enterococcus faecium and a detection method using the kit. The kit detects a primer pair of Listeria monocytogenes hlyA and a primer pair of the dd1 gene of an Enterococcus faecium strain; if specific amplification bands are obtained at 174 bp and 557 bp, Listeria monocytogenes and Enterococcus faecium are positive; and the kit can simultaneously detect Listeria monocytogenes and Enterococcus faecium. The lower limit of the detection sensitivity of the is lower than a minimum infective dose of pathogenic bacteria, which is of great significance to later clinical application and to monitoring of Listeria monocytogenes and Enterococcus faecium.
Owner:INNER MONGOLIA AUTONOMOUS REGION ACAD OF AGRI & ANIMAL HUSBANDRY SCI
Method for breeding rape self-incompatible hybrid seeds under assistance of S haplotype molecular markers
InactiveCN109588306ABreed fastShorten the breeding cycleMicrobiological testing/measurementPlant genotype modificationDuplex pcrHybrid seed
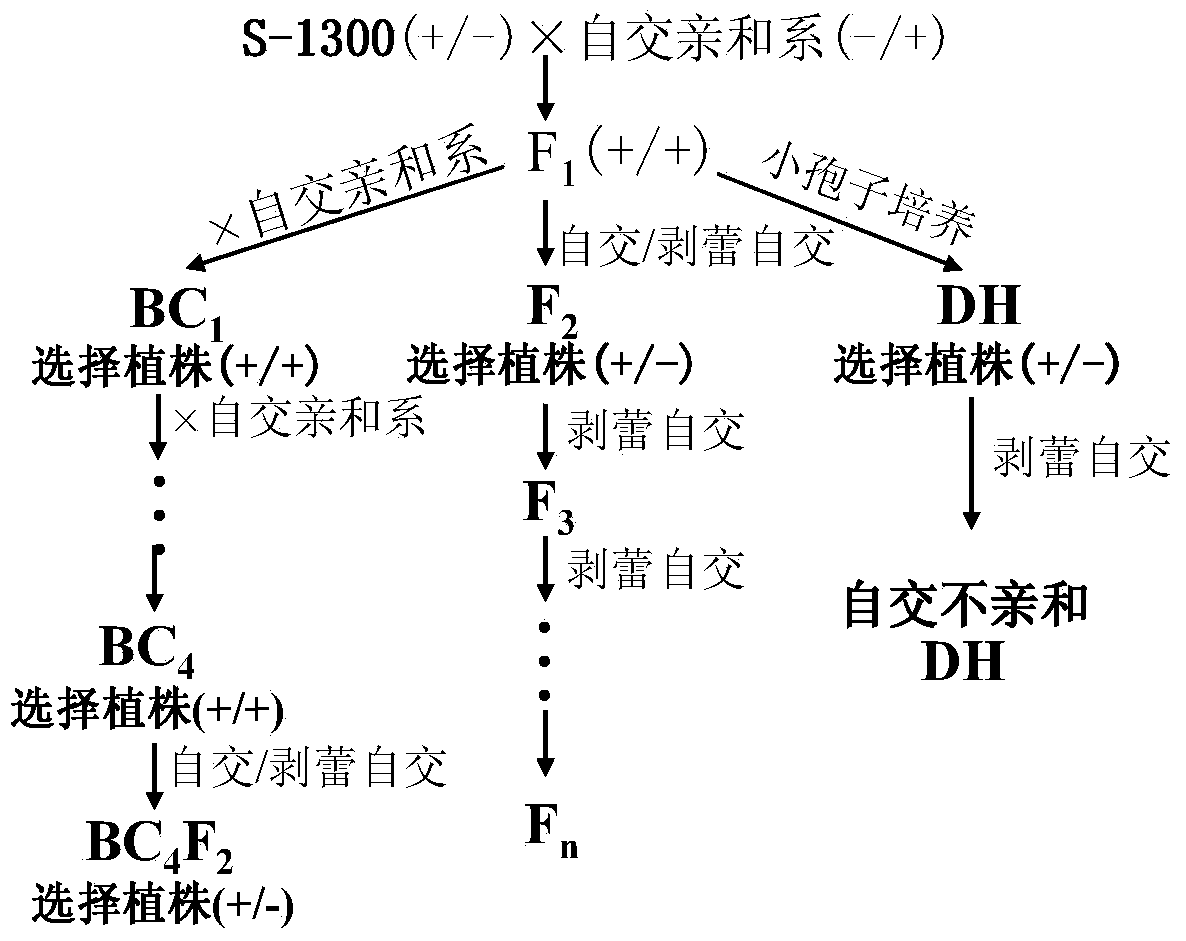
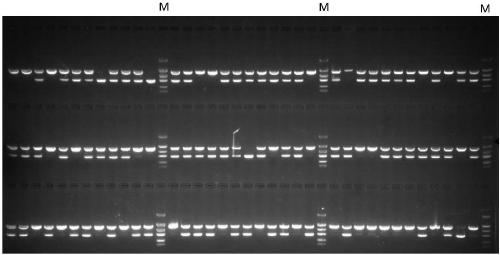
The invention belongs to the technical field of rape molecular breeding, and discloses a method for breeding rape self-incompatible hybrid seeds under assistance of S haplotype molecular markers. Themethod comprises the following steps: obtaining F1 seeds, detecting S haplotype of a selfing line, obtaining self-incompatible two-line and three-line hybrid seeds, and arranging a variety screening test by the obtained two-line hybrid seeds and three-line hybrid seeds. The S haplotype of F1 is identified by double PCR markers, the technical defect that no marker is generated due to accidental factors in the experimental process is overcome, and the S haplotype of the selfing line and recovery or retention of the S haplotype can be accurately judged; fussiness of fertility identification and blindness of combination configuration in the breeding process of rape self-incompatible hybrids are overcome with the molecular marker technology, the breeding workload is greatly reduced, the breeding cycle is shortened, breeding efficiency is improved, and rapid cultivation of the cabbage type rape self-incompatible hybrids is promoted.
Owner:HUAZHONG AGRI UNIV
Method for detecting single root knot of enterolobium cyclocarpum meloidogyne and application thereof
ActiveCN102367484AHigh detection sensitivityShorten detection timeMicrobiological testing/measurementForward primerDuplex pcr
The invention discloses a method for detecting a single knot of enterolobium cyclocarpum meloidogyne and application thereof. In the invention, universal primers MF (Forward Primer) and MR (Reverse Primer) for amplifying meloidogyne and specific primers FMe (Forward Primer) and RMe (Reverse Primer) for amplifying the enterolobium cyclocarpum meloidogyne are designed; a single root knot DNA containing the meloidogyne is taken as a template for PCR (Polymerase Chain Reaction) amplification reaction; a PCR amplification reaction product is subjected to lipolysaccharide electrophoresis detection,and a detection result is obtained according to the existence / absence and the size of straps; and the sequences of the primers are respectively shown as SEQ ID NO:1, SEQ ID NO:2, SEQ ID NO:3 and SEQ ID NO:4 in a table. The one-step dual PCR detection method disclosed by the invention has the advantages of increased molecular detection speed, increased sensitivity and accuracy, and has a practicalapplication value in the aspect of detecting the enterolobium cyclocarpum meloidogyne.
Owner:嘉兴卓十生物科技有限公司
Detection method of early molecule of radopholus similes thorne
InactiveCN101792755AEfficient extractionEarly detectionMicrobiological testing/measurementDNA preparationDuplex pcrDisease
The invention discloses a detection method of an early molecule of radopholus similes thorne, which belongs to the biotechnical field and relates to a radopholus similes thorne DNA extraction solution formula and an extraction method in a morbidity plant tissue, specific primers RsF1 / RsR1 sequences of the radopholus similes thorne, a formula of a one-step double polymerase chain reaction (PCR) reaction solution for combining specific primers of the radopholus similes thorne and primers of D3A and D3B as well as a detection technical method of a rapid specific amplification procedure. The detection method is rapid and accurate while improving the detection sensitivity of the molecule and has high application value in the aspects of the detection on the radopholus similes thorne and early diagnosis of radopholus similes thorne disease.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
Double PCR primer, double PCR detection method and double PCR detection kit for grouper iridovirus
InactiveCN107988433AIncreased sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesDuplex pcrSynchronous detection
The invention provides a double PCR primer, a double PCR detection method and a double PCR detection kit for grouper iridovirus. For the kit provided by the invention, the double PCR primer is designed according to major capsid protein (MCP) gene sequences of the genus megalocytivirus of grouper iridovirus (GIV-M) and the genus ranavirus of grouper iridovirus (GIV-R), a method for synchronously detecting GIV-M and GIV-R through a single tube is established, an amplification system and a method are optimized, and the high-specificity, high-sensitivity and rapid synchronous detection for GIV-M and GIV-R is finally realized.
Owner:SOUTH CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Duplex PCR method for identifying genetic relationship of black carp
ActiveCN109468391ARealize genetic diversity detectionAchieve individual identificationMicrobiological testing/measurementDNA/RNA fragmentationDuplex pcrElectrophoresis
The invention provides a duplex PCR method for identifying the genetic relationship of black carp. The method comprises the steps as follows: (1) extracting genome DNA of a black carp sample; (2) performing duplex PCR by 5 groups of microsatellite primers; (3) performing electrophoretic separation on a PCR amplified product on polyacrylamide gel; (4) counting genotype of the PCR amplified productand analyzing the genetic relationship between individuals. The 5 groups of microsatellite primers are sites with high polymorphism and clear strips and can realize genetic diversity detection and individual recognition of the black carp and identify the genetic relationship between individuals, inbreeding among black carp individuals can be avoided, and a the used time and cost are a half of those in traditional PCRs. Tests prove that the method can identify the genetic relationship of 30 black carps and accurately identify the genetic relationship between samples, and the genetic distances between the samples can be calculated according to identification results.
Owner:YANGTZE RIVER FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Dual PCR method for simultaneously detecting Listeria monocytogene and Listeria ivanovii
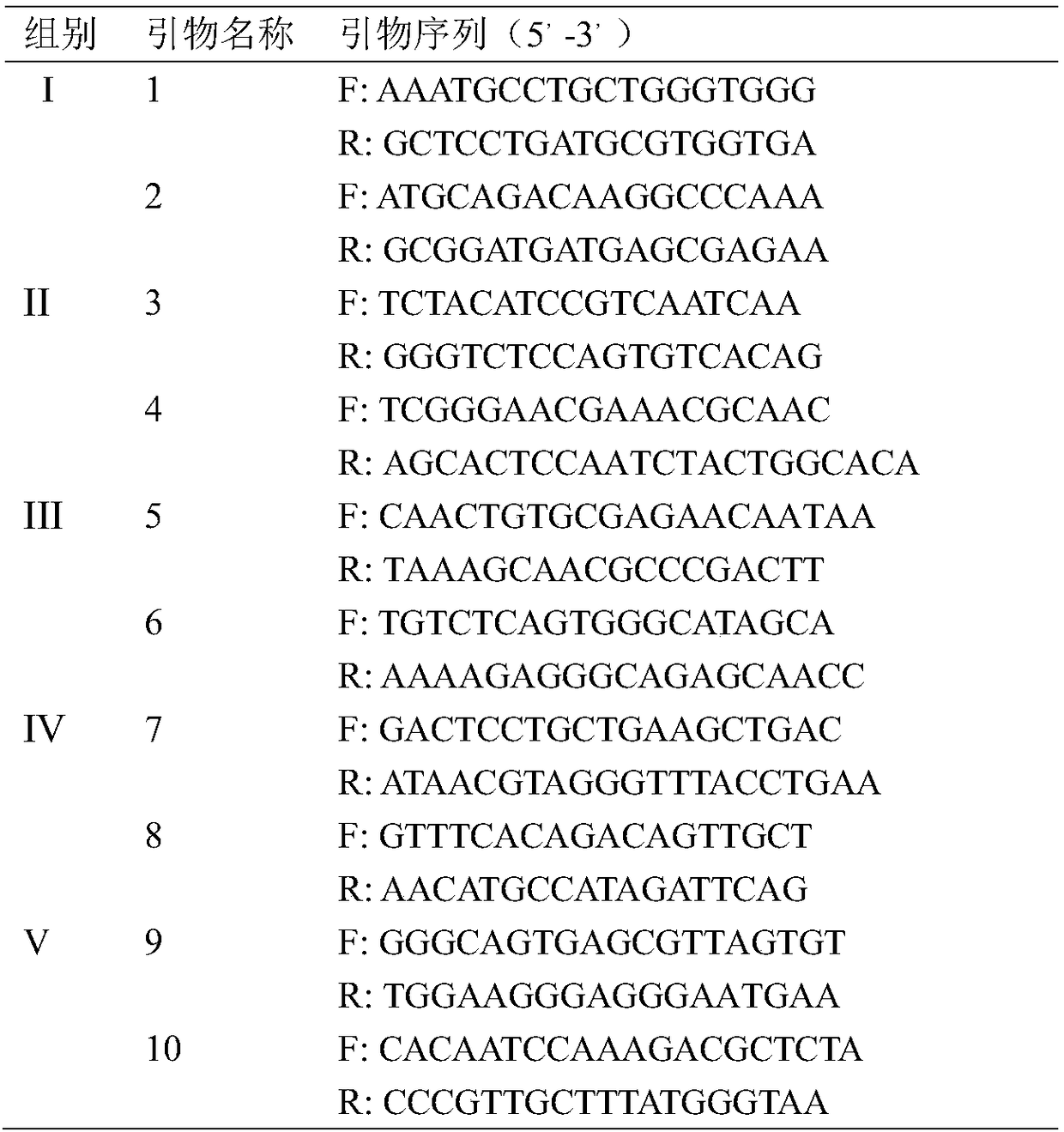
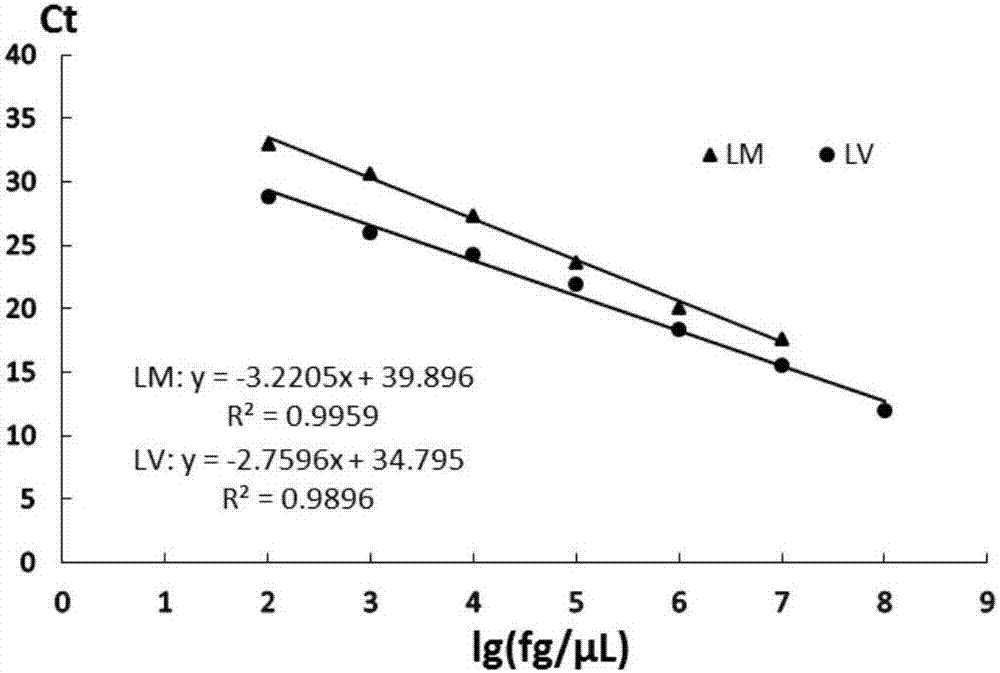
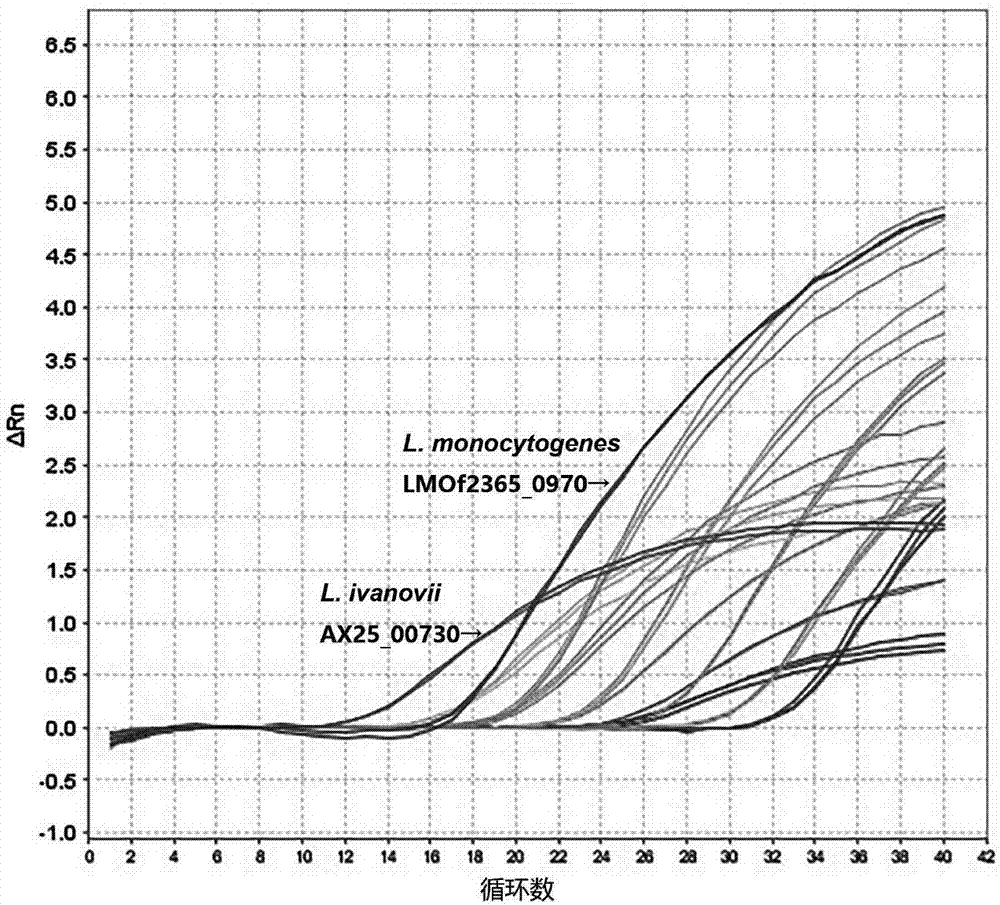
InactiveCN107988330AStrong specificityEfficient detectionMicrobiological testing/measurementMicroorganism based processesDuplex pcrFluorescence
The invention discloses a dual PCR method for simultaneously detecting Listeria monocytogene and Listeria ivanovii, and belongs to the technical field of detection of foodborne pathogenic bacteria. Primers and probes are designed in allusion to a newly discovered Listeria monocytogene-specific gene LMOf2365_0970 and a newly discovered Listeria ivanovii-specific gene AX25_00730; by the detection method, the Listeria monocytogene and the Listeria ivanovii in food can be effectively detected; the detection limit for the two target bacteria is N*102 CFU / mL, and the fluorescence quantitative PCR detection time is shorter than 40 minutes; the detection method has the characteristics of high detection sensitivity, good specificity and strong anti-interference ability; by the detection method, quick qualitative and accurate quantitative detection of the Listeria monocytogene and the Listeria ivanovii in food can be achieved, and the purpose of rapidly screening contaminated food samples is achieved.
Owner:NANJING AGRICULTURAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com