Duplex PCR method for detecting porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus
A porcine epidemic diarrhea and infectious technology, which is applied in biochemical equipment and methods, microbial determination/examination, etc., can solve the problems of inappropriate epidemiological investigation, time-consuming and laborious, inappropriate clinical rapid diagnosis, etc., and achieves high sensitivity and detection. Efficient and specific effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] 1 RNA extraction
[0050] The method is the same as in Comparative Example 1.
[0051] 2 cDNA synthesis
[0052] The method is the same as in Comparative Example 1.
[0053] 3 Establishment of Multiplex PCR
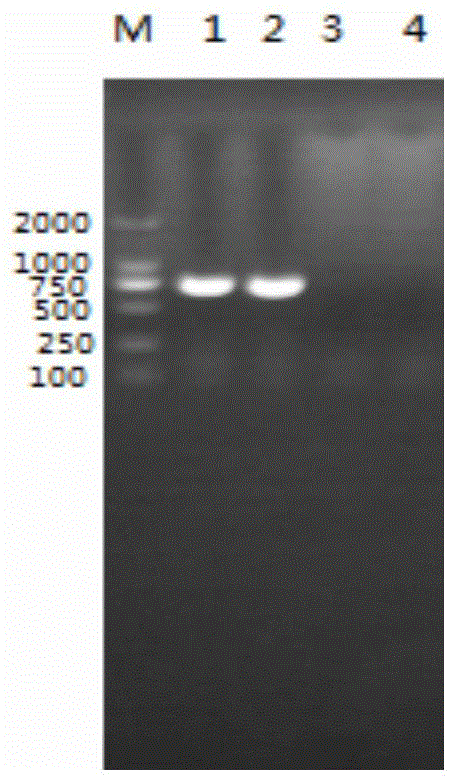
[0054] RNA was extracted from samples suspected of being infected with TGEV or (and) PEDV, and cDNA was transcribed to obtain cDNA. Using this cDNA as a template, the sequences were respectively SEQ ID NO.1, SEQ ID NO.2, SEQ ID NO.3, and SEQ ID NO. The 2 pairs of primers in 4 were added to the amplification system at the same time to establish a double PCR amplification reaction system. The double PCR reaction system is as follows: 1 μl of cDNA template, 0.5 μl of 2 pairs of upstream and downstream primers, 12.5 μl of PCR Mix, and 50 μl of ultrapure water. PCR reaction conditions: pre-denature at 95°C for 5 minutes, enter a cycle of 95°C for 60 seconds, 52°C for 1 minute, and 72°C for 1 minute, cycle 35 times, and finally extend at 72°C for 5 minutes. After PCR...
Embodiment 2
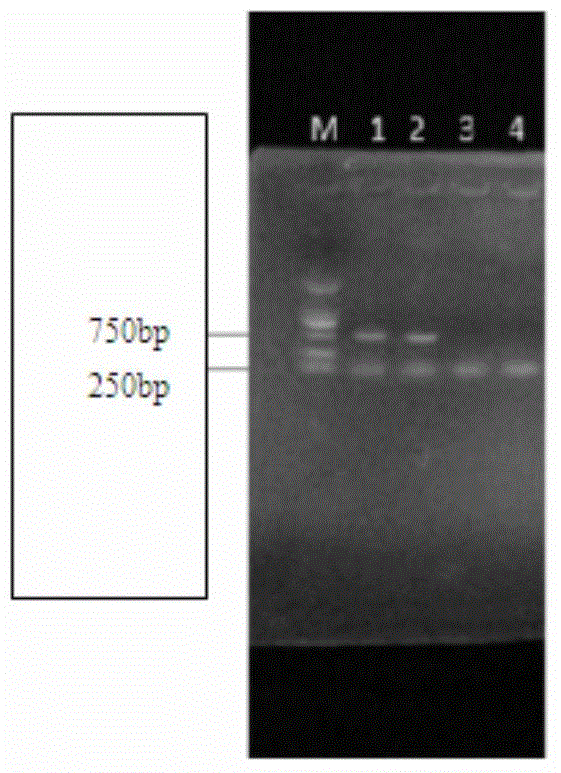
[0057] Example 2 Verifies the specificity experiment of multiplex PCR.
[0058] 1 RNA extraction
[0059] The method is the same as in Comparative Example 1.
[0060] 2 cDNA synthesis
[0061] The method is the same as that in Comparative Example 1. The RNA of porcine respiratory and reproductive syndrome virus (PRRSV), classical swine fever virus (CSFV) and porcine rotavirus (PRAV) is extracted and reverse-transcribed to obtain cDNA, which is carried out with the cDNA templates of PEDV and TGEV. Multiplex RT-PCR reactions.
[0062] 3 Multiplex PCR detection
[0063] Using the cDNA synthesized in step 2 as a template, add two pairs of primers whose sequences are SEQ ID NO.1, SEQ ID NO.2, SEQ ID NO.3, and SEQ ID NO.4 to the amplification system at the same time to establish a double PCR Amplification reaction system. The double PCR reaction system was as follows: 1 μl of cDNA template, 0.5 μl of each pair of upstream and downstream primers, 12.5 μl of PCRMix, and 50 μl of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com