Identification method for determining anastomosis groups of rhizoctonia solani
A technology of a rice sheath blight fungus and an identification method, which is applied in the identification field of the determination of the fusion group of the rice sheath blight fungus, and can solve the problems of inability to distinguish multiple fusion group types, disadvantage, insufficient rice sheath blight fungus, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Isolation and purification of rice sheath blight from susceptible rice stalks
[0044] The rice stems of Guodao No. 6 hybrid rice susceptible to sheath blight were collected from the rice fields of the Fuyang Experimental Base of the China Rice Research Institute and brought back to the laboratory. Rinse the diseased stem tissue three times with sterile water, and blot the moisture on the surface of the tissue with sterilized filter paper. Then cut the diseased tissue into small pieces with a length and width of 5 mm, place it in 2% water agar (20 g of agar, 1000 mL of water) medium, and culture it in an incubator at 28 ° C. After 24 to 36 hours, silk grows around the diseased tissue. The colony with Sclerotinia characteristics was cut out with a sterile blade and transplanted into a PDA (200g potato, 20g glucose, 20g agar, 1000mL water) medium for purification. After purification and culture in an incubator at 28°C, it was stored in Store in the refrigerator...
Embodiment 2
[0045] Example 2 Extraction of bacterial strain genomic DNA
[0046] Acquisition of mycelium: After activation of the standard strain of Rhizoctonia solani or isolated rice sheath blight bacteria preserved on the slant medium, pick a small piece of mycelium (about 0.5mm×0.5mm) Transfer to a 250mL Erlenmeyer flask filled with 100mL PDB (200g potato, 20g glucose, 1000mL water) medium, and shake and culture at 28°C for 5-6d. The obtained culture was rinsed twice with sterilized water, then the culture was fully dried with a vacuum pump, and stored in a -20°C refrigerator for later use.
[0047] DNA extraction: The total genomic DNA of the strains was extracted by the improved CTAB method. Take 0.2 g of blotted hyphae, grind it with liquid nitrogen, add 800 μL DNA extraction solution (1L solution contains 20 g CTAB, 1.4 M NaCl, 100 mM Tris-HCl, 20 mM EDTA and 1 ml β-mercaptoethanol, pH value is 8.0), in Incubate in a 1.5mL centrifuge tube at 65°C for 30min, shake gently 3-4 time...
Embodiment 3
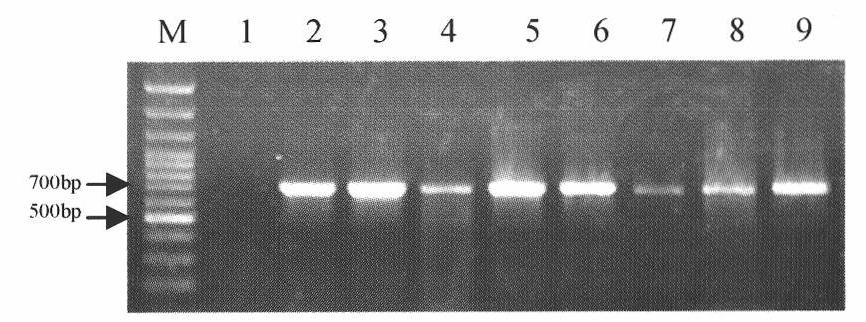
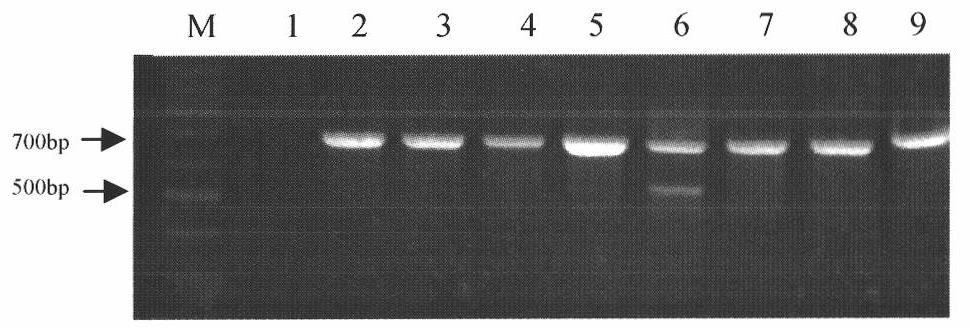
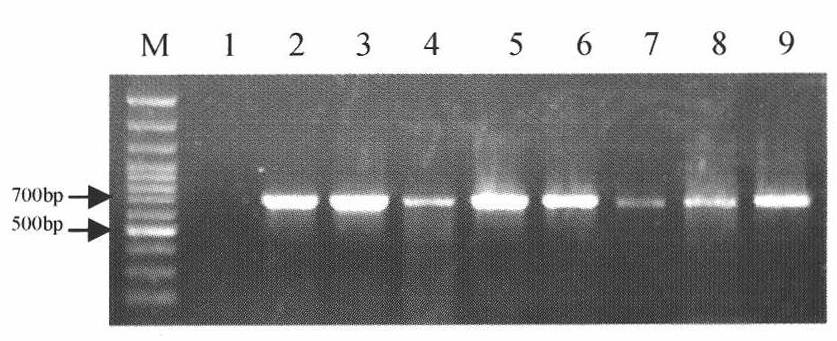
[0051] Example 3 PCR Amplification of the ITS-5.8S rDNA Sequence of Different Mycelia Fusion Group Standard Strains
[0052] For the standard strains R1-R8 belonging to different hyphae fusion group strains AG-1IA, AG-1IB, AG-2-1, AG-4, AG-5, AG-6, AG-8 or AG-BI, carry out PCR amplification of ITS-5.8S rDNA sequence. Universal primers ITS1 and ITS4 designed by White et al. (1990) were synthesized by Shanghai Bioengineering Co., Ltd. The sequences of ITS1 and ITS4 are: ITS1 (5'-TCCGTAGGTGAACCTGCGG-3'; SEQ No.1), ITS4 (5' - TCCTCCGCTTATTGATATGC-3'; SEQ No. 2). The PCR reaction system is: 2.5 μL of 10×Taq reaction buffer in a total volume of 25 μL, 25 mM MgCl 2 , 2.5mMdNTP, 10μM primer ITS1, 10μM primer ITS4, standard strain DNA 100ng, 0.5U Taq DNApolymerase, add ddH 2 O to make up the volume to 25 μL. The PCR reaction conditions were: pre-denaturation at 94°C for 4 minutes; denaturation at 94°C for 50 s, annealing at 55°C for 50 s, and extension at 72°C for 50 s, a total of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com