Dual PCR method for simultaneously detecting Listeria monocytogene and Listeria ivanovii
A technology for Listeria monocytogenes and Listeria yiseri, which is applied in the directions of microorganism-based methods, biochemical equipment and methods, and microorganism determination/inspection, etc. Good specificity and high detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Establishment of a dual real-time fluorescent quantitative PCR detection system for Listeria monocytogenes and Listeria ishii
[0030] Through the comparison of the whole genome sequences of Listeria monocytogenes and Listeria eziri, two species-specific genes were screened: LMOf2365_0970 (YP_013571.1) and AX25_00730 (AHI54712.1), and the gene LMOf2365_0970 was only found in single 100% homology within Listeria species, gene AX25_00730 is 100% homologous only to Listeria iwiri. Beacon Designer 8.0 software was used to design specific primers and probes, and blastn comparison analysis was carried out in the NCBI database, and then the PCR amplification results of the primers to be selected were simulated through the online bioinformatics tool "in silico". PCR amplification was used to verify the specificity of the primers, and the sequences of the fluorescent quantitative PCR primers were confirmed as follows:
[0031] TQM-f: 5'-cagaccgttatagtgaattc-3'. (SEQ...
Embodiment 2
[0043] Embodiment 2: Specificity evaluation of the dual real-time fluorescent quantitative PCR detection system of the present invention
[0044] Inoculate 36 strains of Listeria monocytogenes (including 13 serotypes), 1 strain of Listeria izini and 5 strains of other Listeria in the modified Listeria enrichment medium, and shake and cultivate at 37°C for 10 hours at 180rpm; Take 1 mL of the bacterial liquid in a 1.5 mL centrifuge tube, extract and purify the genomic DNA according to the conventional genome extraction method; the genomic DNA is stored at -20°C for later use. Among them, the components of the improved Listeria enrichment medium are: tryptone 17.0g / L, polyvalent peptone 3.0g / L, sodium chloride 5.0g / L, potassium dihydrogen phosphate 1.4g / L, disodium hydrogen phosphate 12.0 g / L, escin 1.0g / L, acriflavine 0.02g / L, nalidixic acid 0.018g / L, lithium chloride 0.015g / L, sodium pyruvate 0.7g / L, dissolved in 1000mL distilled water middle. ), the genomic DNA extraction a...
Embodiment 3
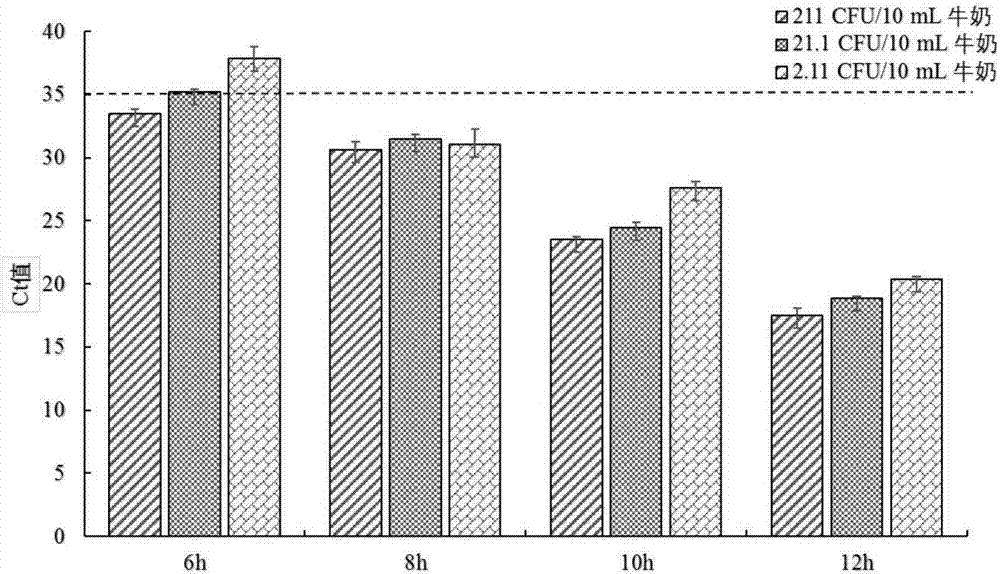
[0050] Example 3: Sensitivity, anti-interference and repeatability evaluation of dual real-time fluorescent quantitative PCR detection system
[0051] (1) Sensitivity evaluation
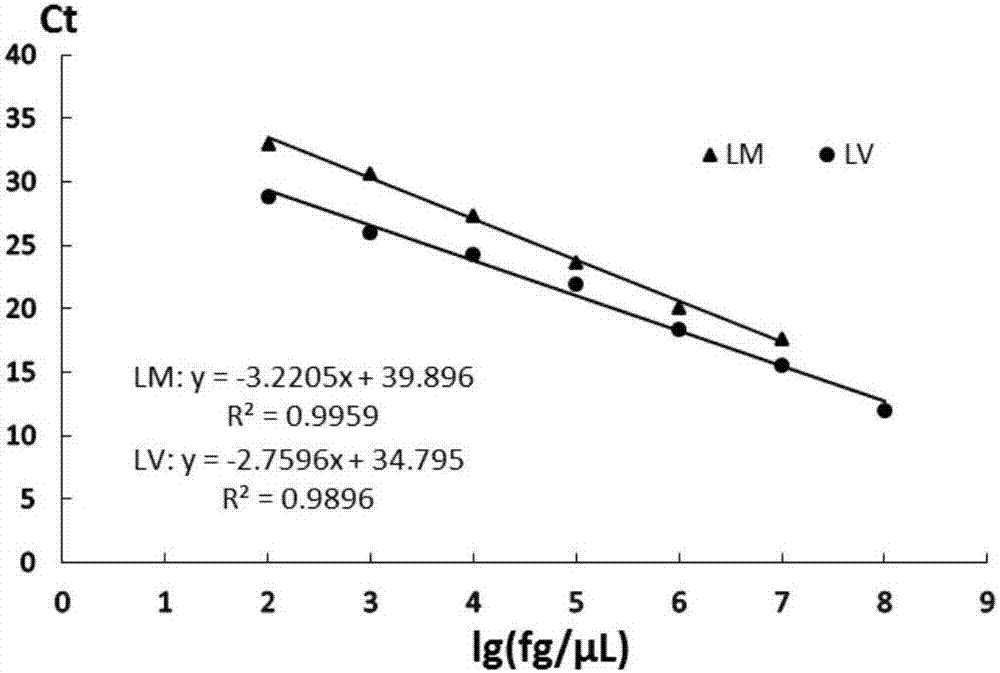
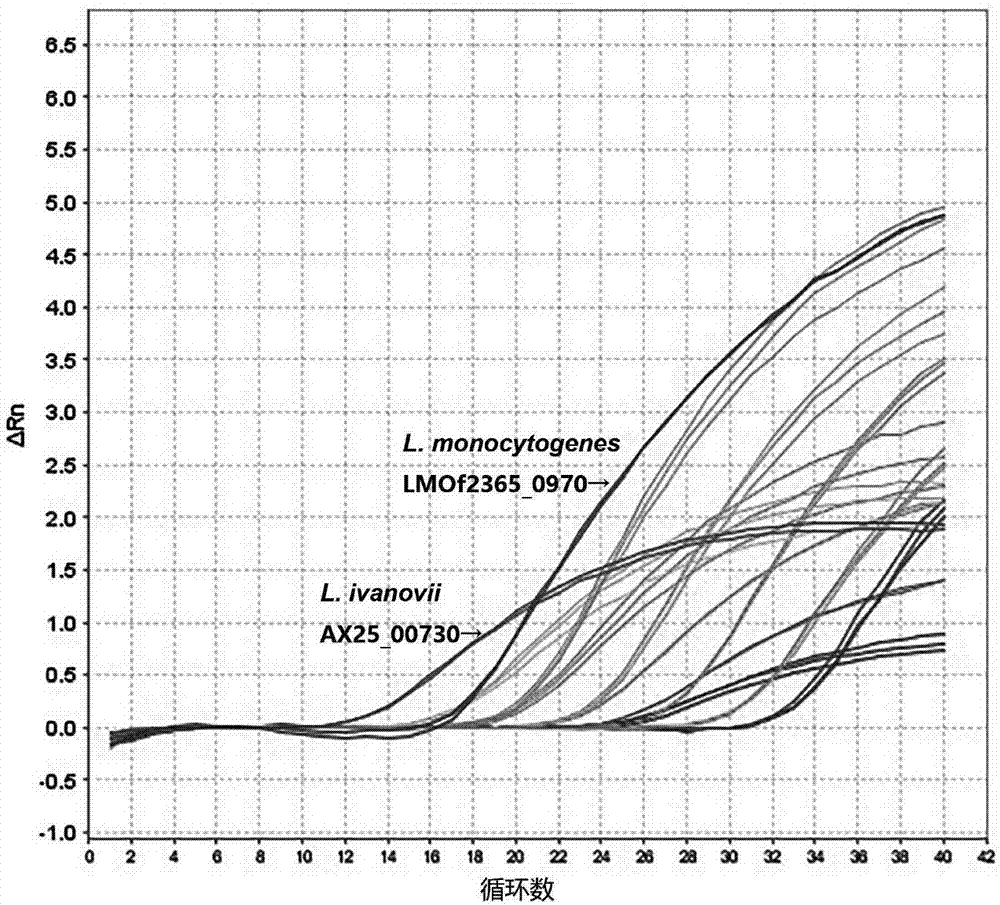
[0052] The genomic DNA of Listeria monocytogenes and Listeria eziri were serially diluted 10 times with TE buffer respectively, and the initial concentrations of the genomic DNA templates of the two bacteria were measured by an ultra-micro nucleic acid analyzer, which were 61.6ng / μL and 187.1ng / μL. 2 μL of DNA in each dilution gradient was used as a template for real-time fluorescent quantitative PCR reaction, and three parallel samples were made for each dilution gradient, and sterile water was used as a blank control. The results are shown in Table 2 and attached figure 2 As shown, when the concentration of Listeria monocytogenes is in the range of 61.6ng / μL~61.6fg / μL, the primer TQM can obtain credible positive results, and when the DNA concentration drops to 6.16fg / μL, the reaction system Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com