Method for detecting single root knot of enterolobium cyclocarpum meloidogyne and application thereof
A root-knot nematode and detection method technology, applied in the biological field, can solve the problem of accurate identification of template DNA from nematodes and root-knot nematodes, achieve high practical application value, reduce detection time, and improve sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] (1) Obtaining single knot DNA
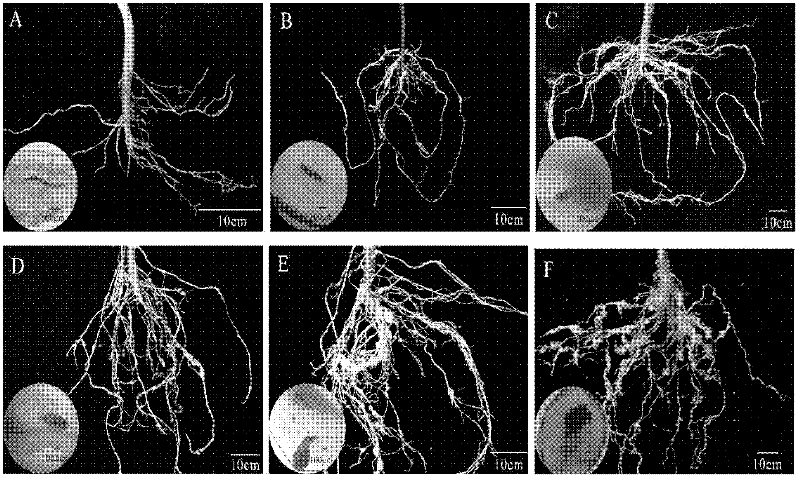
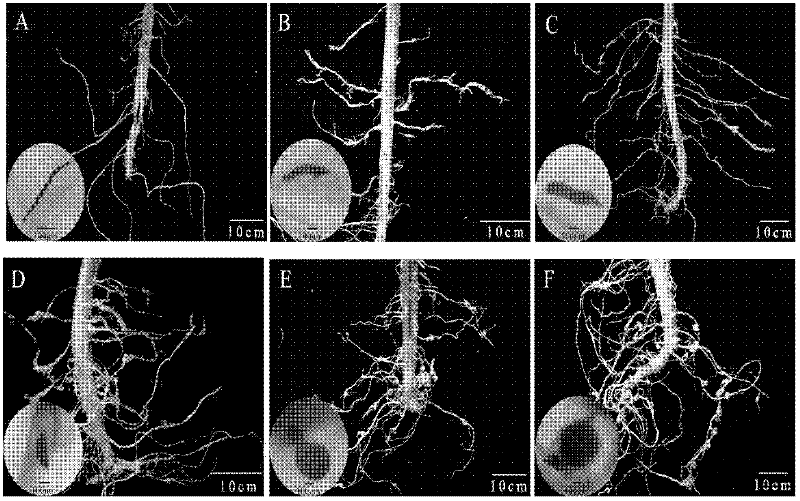
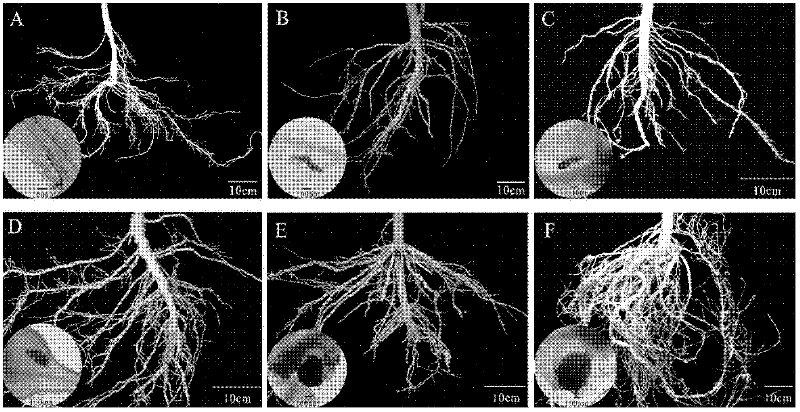
[0039] Sow tomato (Xiahong No. 1), cucumber (Xinwanji) and water spinach 25 days later and transplant the seedlings to flower pots with sterilized soil with a diameter of 15 cm, 1 plant per pot. One week after transplanting the seedlings, each plant was inoculated with 200 2nd instar larvae (J2) of Meloidogyne elegans, Meloidogyne incognita, Meloidogyne javanica and Root-knot nematode peanut, while keeping healthy plants that were not inoculated as controls , placed in a light culture room for 14 hours of light and 10 hours of darkness for constant temperature cultivation at 25°C. The morphology of nematodes was observed by staining at 5d, 10d, 15d, 20d, 25d, and 30d after inoculation, respectively. At the same time, healthy root tissue DNA and single root junction DNA were extracted, and 5 replicates were taken for each treatment, and 1 blank control was set.
[0040] Root tissue staining and nematode morphology observation at differen...
Embodiment 2
[0050] (2) Synthetic primers
[0051] 根据植物线虫大亚基核糖体DNA(基因登录号为:AF435803、AF435794、AY446970、JN005857、FN429017、GQ375158、EU364890、EU570214、AF435793、AF435802、FJ485651、DQ328713、HQ688681、EU130893和EU36859)的D2D3区设计线虫 通用引物MF和MR;根据根结线虫mtDNA COII和lrRNA基因(基因登录号为:FJ159631、AY635612、GQ266686、GQ266685、AY446970、GQ870255、AY757882、GQ865513、HM161680、AY635609、AY757909、AY942848、AY942851和EU364883)间 The specific primers FMe and Rme of Meloidogyne elegans were designed based on regional interspecific differences.
[0052] The nucleotide sequence of the primer is as follows:
[0053] MF 5'-GGGGATGTTTGAGGCAGATTTGT-3';
[0054] MR 5'-AACCGCTTCGGACTTCCACCAG-3';
[0055] FMe 5'-CATTCATTTATACCAATTTTAGTTGAGG-3';
[0056] RMe 5'-CAATTATTGAATATTTTTTTCCCAAACGA-3'.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com