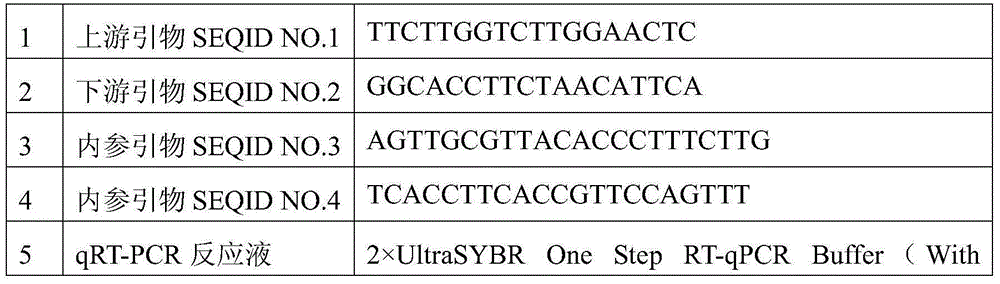
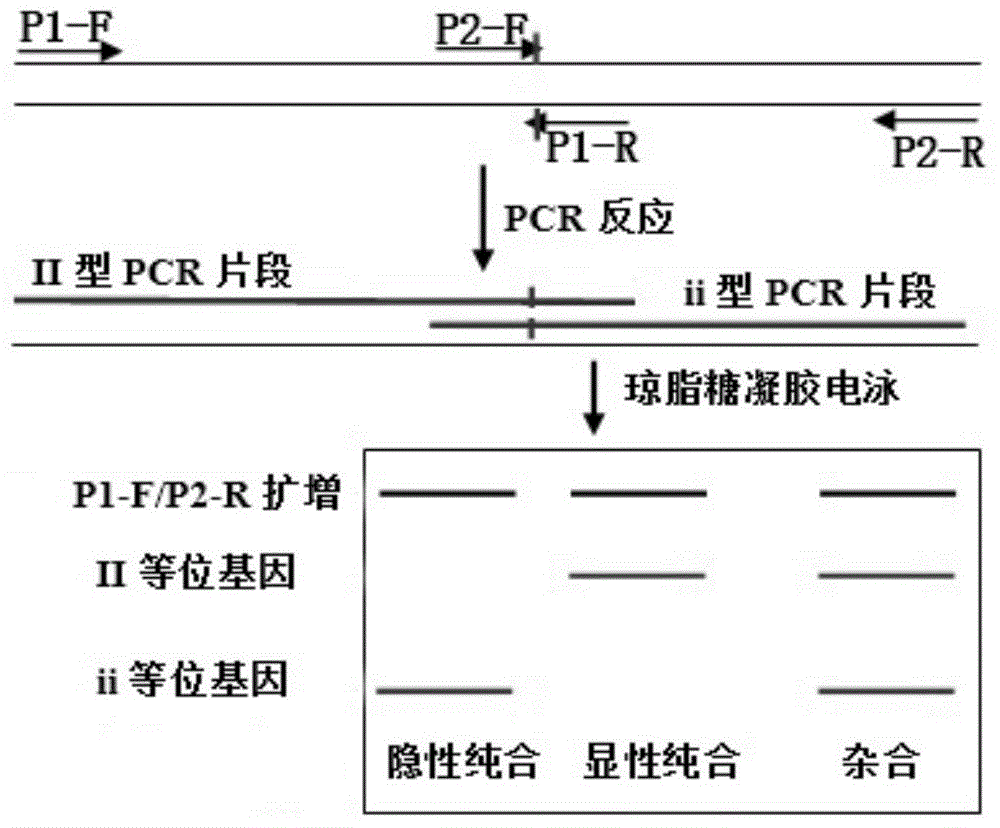
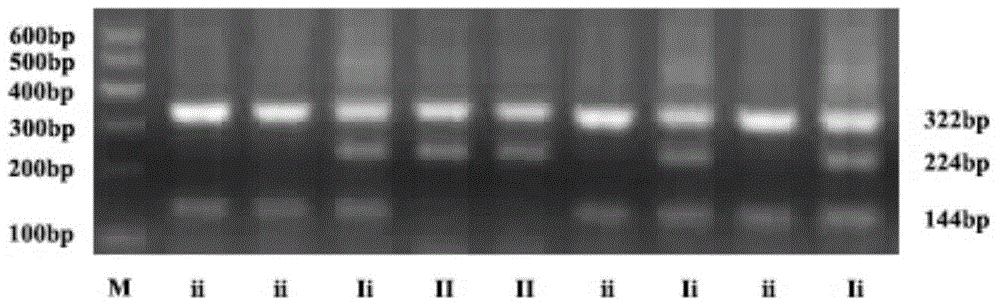
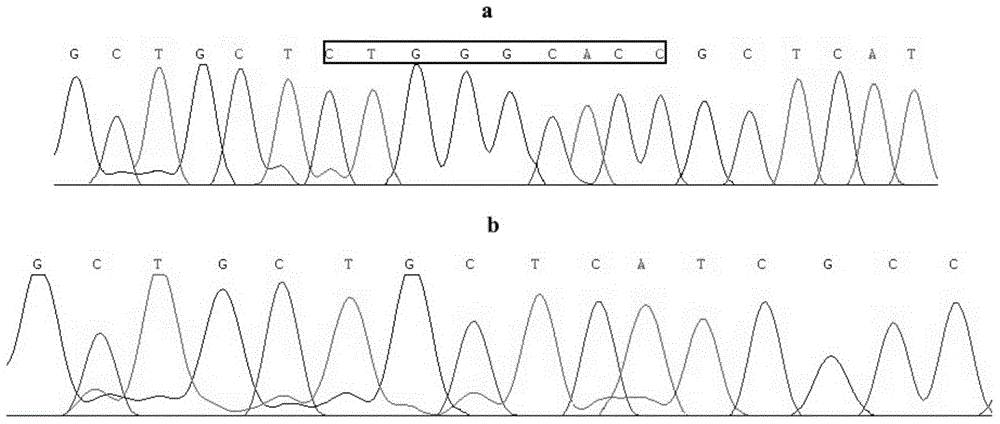
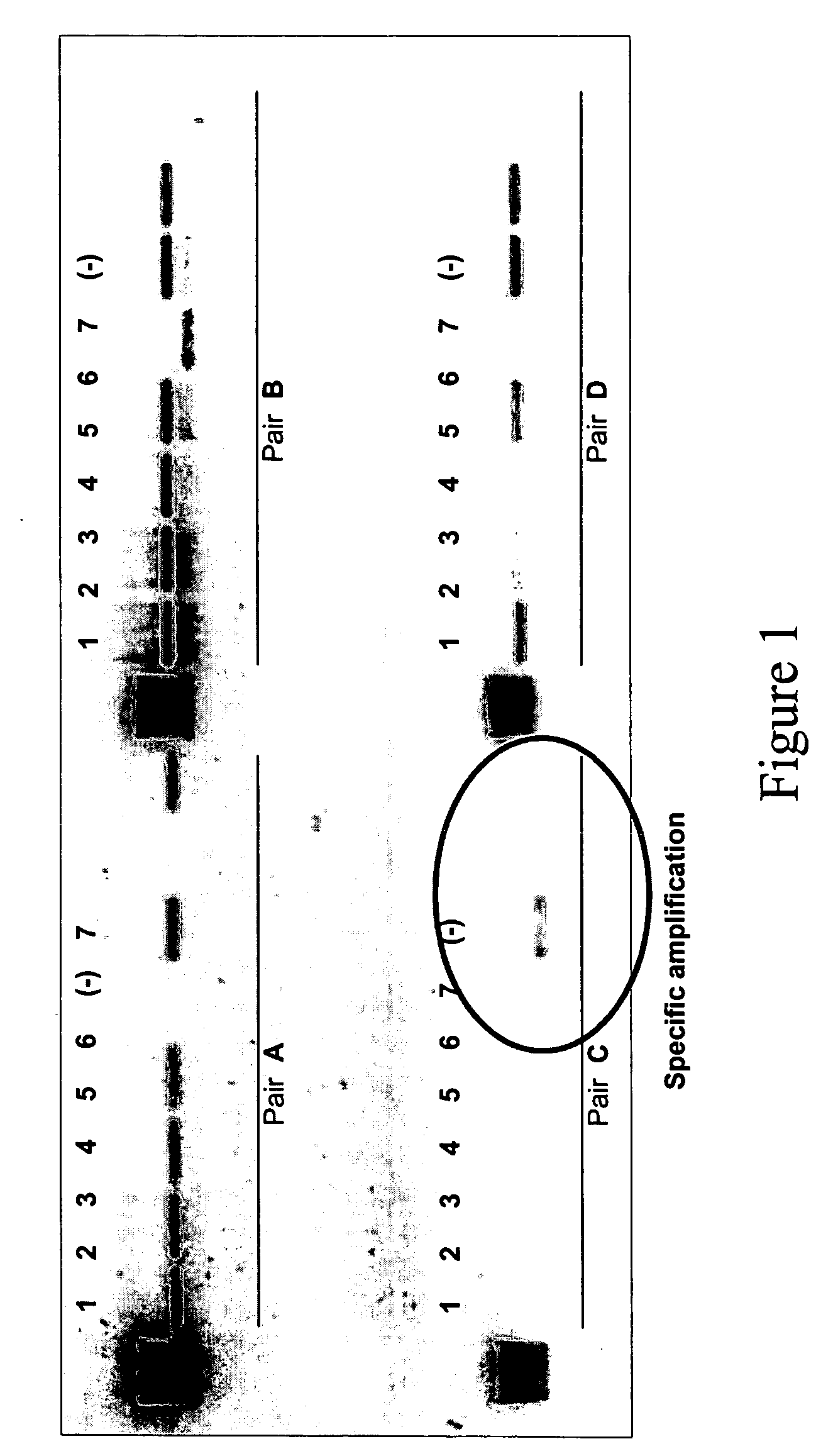
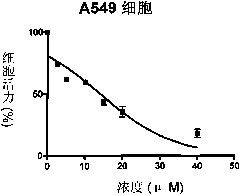
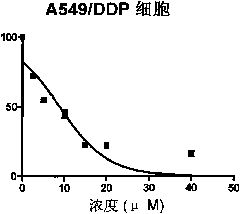
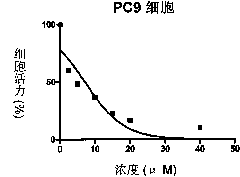
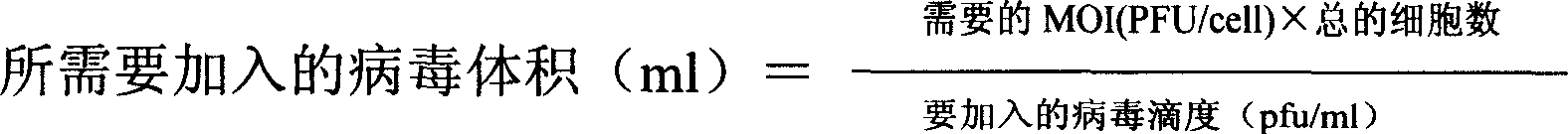Patents
Literature
370 results about "MOLECULAR BIOLOGY METHODS" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
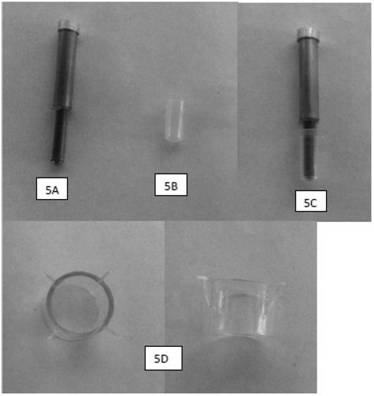
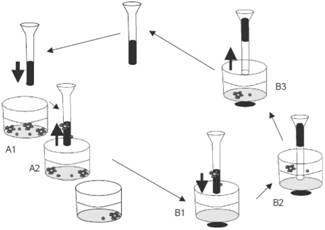
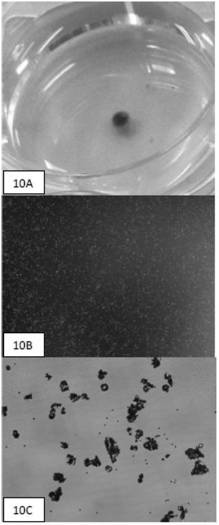
Cell enriching, separating and extracting method and instrument and single cell analysis method
ActiveCN102690786AEnrichment reachedEfficient collectionMicrobiological testing/measurementBiomass after-treatmentImmunofluorescence stainingMagnetic bead
The invention discloses a method and an automatic instrument device for enriching and extracting target cells and separating single cells by using a positive magnetic bead method and performing immunity and molecular biology identification and analysis on single cells. By the method and the instrument device, the on / off of a capture magnet and a release magnet is controlled by various methods to complete target cell searching, capturing, cleaning and releasing operation once or for multiple times, so that the target cell detection sensitivity and stability are improved. When the capture magnet searches and captures the target cells, the search line is circular, square, comb-shaped, S-shaped or U-shaped. In addition, the captured substances are filtered, most free micro magnetic beads are removed, the purity of the product is further improved and the product can be used as a good experimental material. By combining a special filter and the adsorption of the capture magnet, more than 95 percent of free micro magnetic beads can be effectively removed. Meanwhile, the types of the single cells are identified and biological characteristics of the single cells are analyzed by an immunofluorescence staining method and a method in molecular biology, and effective biological indexes are provided for clinical diagnosis and treatment of cancers.
Owner:GD TECH INC
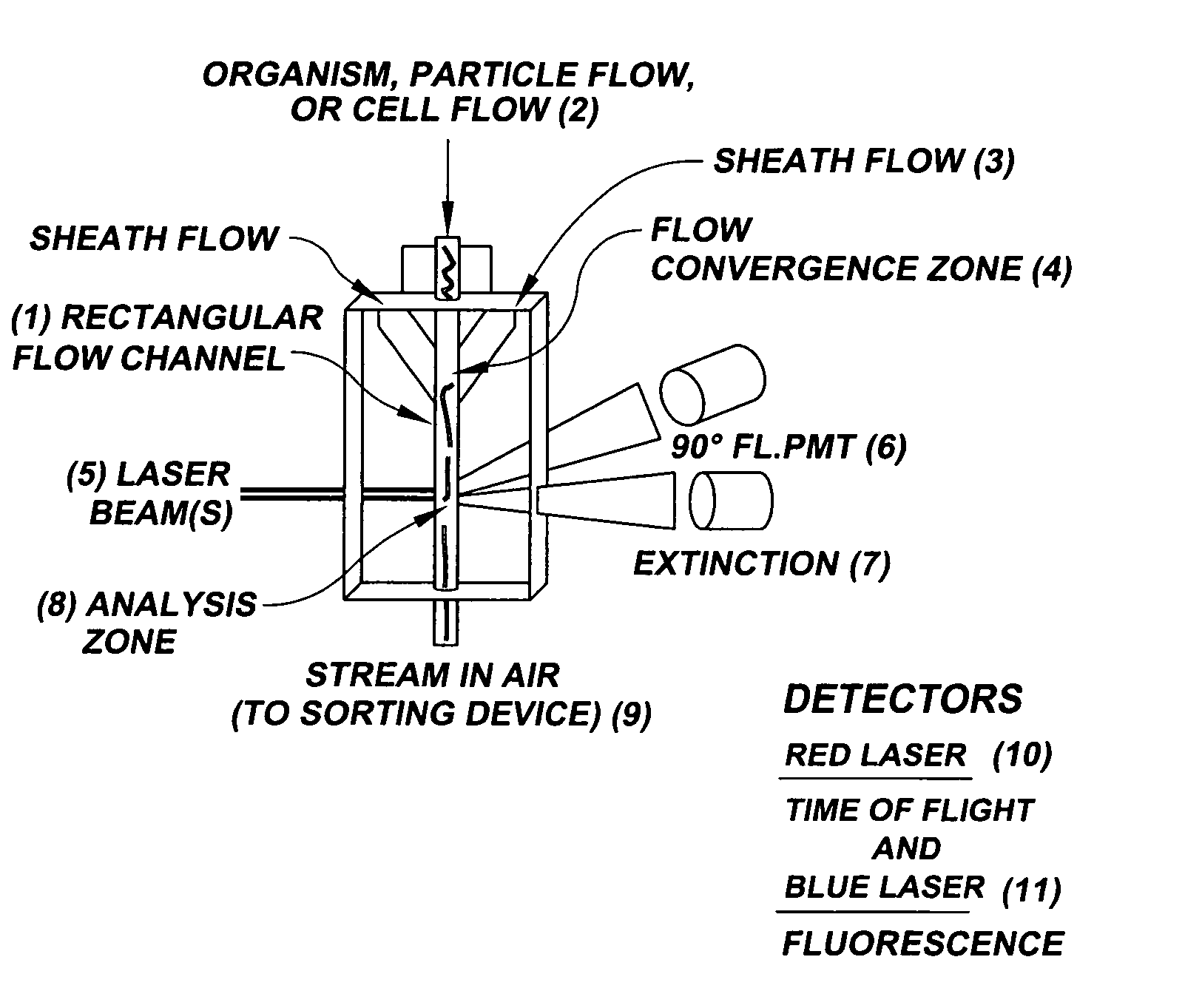
System for axial pattern analysis of multicellular organisms
A method of using elongate multicellular organisms in conjunction with a specialized flow cytometer for drug discovery and compound screening. A stable, optically detectable linear marker pattern on each organism is used to construct a longitudinal map of each organism as it passes through the analysis region of the flow cytometer. This pattern is used to limit complex data analysis to particular regions of each organism thereby simplifying and speeding analysis. The longitudinal marker pattern can be used to alter signal detection modes at known regions of the organism to enhance sensitivity and overall detection effectiveness. A repeating pattern can also be used to add a synchronous element to data analysis. The marker patterns are established using known methods of molecular biology to express various indicator molecules. Inherent features of the organism can be rendered detectable to serve as marker patterns.
Owner:UNION BIOMETRICA
Method for Adjusting and Controlling Microbial Enhanced Oil Recovery
InactiveUS20120214713A1Improve performanceEnhanced overall recoveryBacteriaFlushingMetaboliteMicrobial enhanced oil recovery
The invention discloses a method for adjusting and controlling microbial enhanced oil recovery, which comprises the following steps: (1) analyzing the microbial community structure in produced fluid of an oil reservoir via molecular biological method and / or detecting the metabolites in the produced fluid; (2) adjusting the microorganism(s) to be injected into the oil reservoir and / or the nutrient system corresponding to the microorganism(s); (3) injecting the adjusted microorganism(s) and / or the nutrient system corresponding to the microorganism(s) into the oil reservoir through a water injection well; and (4) obtaining the crude oil from a corresponding beneficial oil producing well. Compared with the prior art, the method of the present invention adjusts microbial community structure in the oil reservoir to evolve toward the direction of facilitating oil production, and the performance of the functional microorganism(s) can be completely realized; the nutrient system is pertinently injected to avoid the blindness of using the nutrient system. Therefore, the method is scientific, economical and effective for the microbial enhanced oil recovery.
Owner:EAST CHINA UNIV OF SCI & TECH
Method for regulating and controlling microbial enhanced oil recovery
ActiveCN101699025AEasy to extract oilOil displacement effect is goodBacteriaMicroorganism based processesBiotechnologyMicrobial oil
The invention relates to a method for regulating and controlling microbial enhanced oil recovery. The method comprises the following steps: (1) analyzing a microbial community structure in oil deposit produced fluid and / or metabolic products in detection produced fluid by adopting a molecular biological method; (2) regulating a microbe to be injected into the oil deposit and / or a nutritious system corresponding to the microbe; (3) injecting the regulated microbe and / or the nutritious system corresponding to the microbe into the oil deposit by a water injection well; and (4) gathering crude oil by a corresponding beneficial producing well. Compared with the prior art, the method for regulating the microbial community structure in the oil deposit contributes to the direction evolution of oil recovery, can sufficiently play the performance of functional microbes, injects the nutritious system specifically, and avoids blindness in application of the nutritious system, thus the method is a scientific, economic and effective microbial oil recovery method.
Owner:DAQING HUALI ENERGY BIOLOGICAL TECH
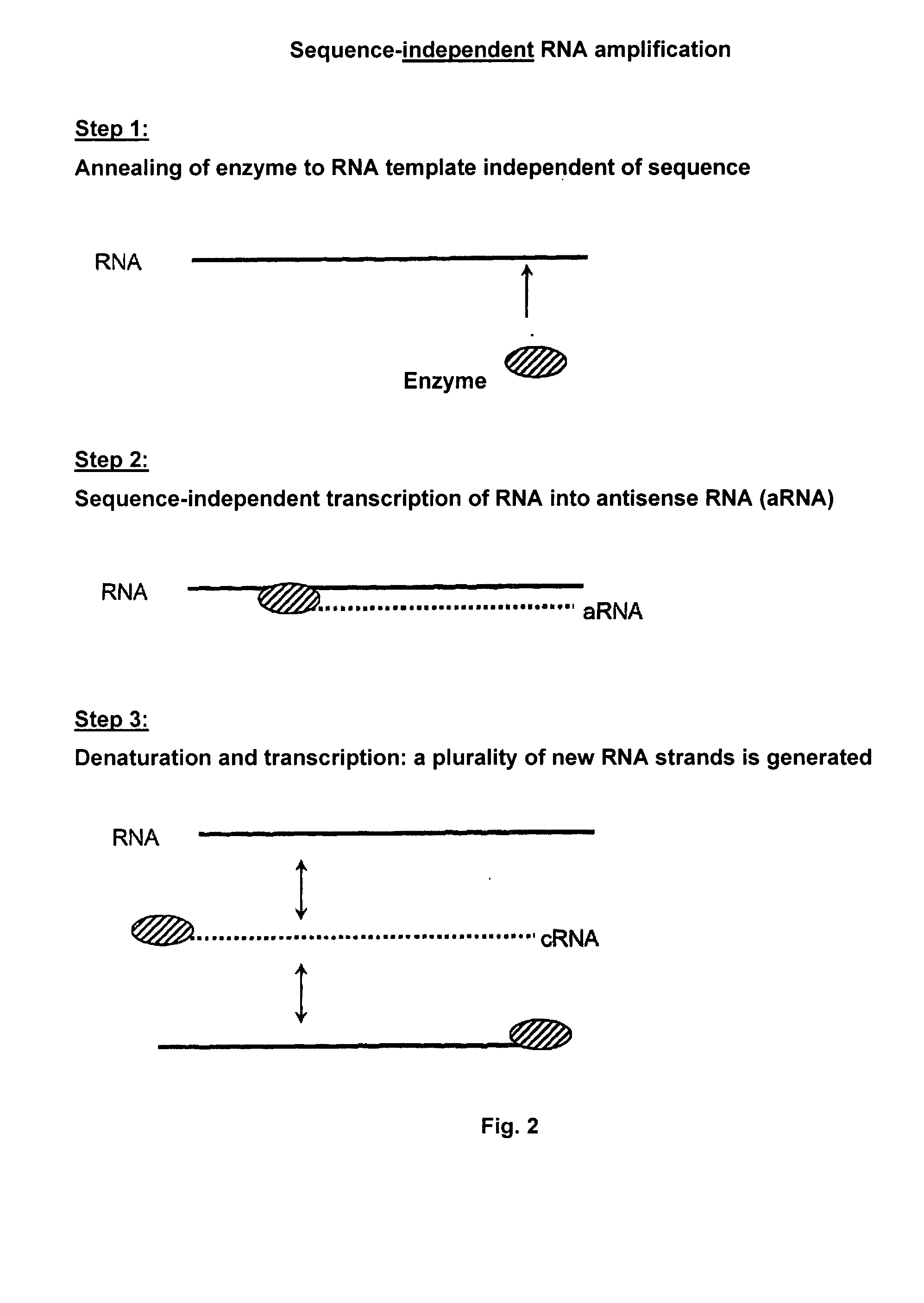
RNA-Dependent RNA Polymerase, Methods And Kits For The Amplification And/Or Labelling Of RNA
InactiveUS20080176293A1Direct and efficient and simple preparationImprove purification effectMicrobiological testing/measurementTransferasesArginineTyrosine
The invention relates to an RNA-dependent RNA-polymerase, to methods and kits for marking and / or amplifying in a primary dependent or independent manner a ribonucleic acid (RNA), in particular a viral, eucaryontic, procaryontic and double-stranded ribonucleic acid (RNA) for the biological and medical use thereof. Said RNA-dependent RNA-polymerase comprises a right-hand conformation and the amino acid sequence the following sequence sections: a. XXDYS b. GXPSG c. YGDD d. XXYGL e. XXXXFLXRXX having the following significances: D: aspartate, Y: tyrosine, S: serine, G: glycine, P: proline, L: leucine, F: phenylalanine, R: arginine, X: any amino acid. The inventive amplifying method is particularly suitable for microarray engineering, siRNA production and for diagnosticating viral infections by detecting viral RNA in patient samples. The inventive marking method is particularly suitable for purifying RNA by means of affinity bonds and for marking RNA used in molecular biology methods for characterising the function and / or structure of a viral, eucaryontic, procaryontic and double-stranded ribonucleic acid (RNA).
Owner:RIBOXX
Mutated DNA polymerases with increased mispairing discrimination
The present invention relates to DNA polymerases with a special mutation which have an enhanced mismatch discrimination, the preparation and use thereof. The thermostable DNA polymerases with this mutation are particularly suitable for diagnostic and molecular-biological methods, e.g., allele-specific PCR.
Owner:UNIVERSITY OF BONN
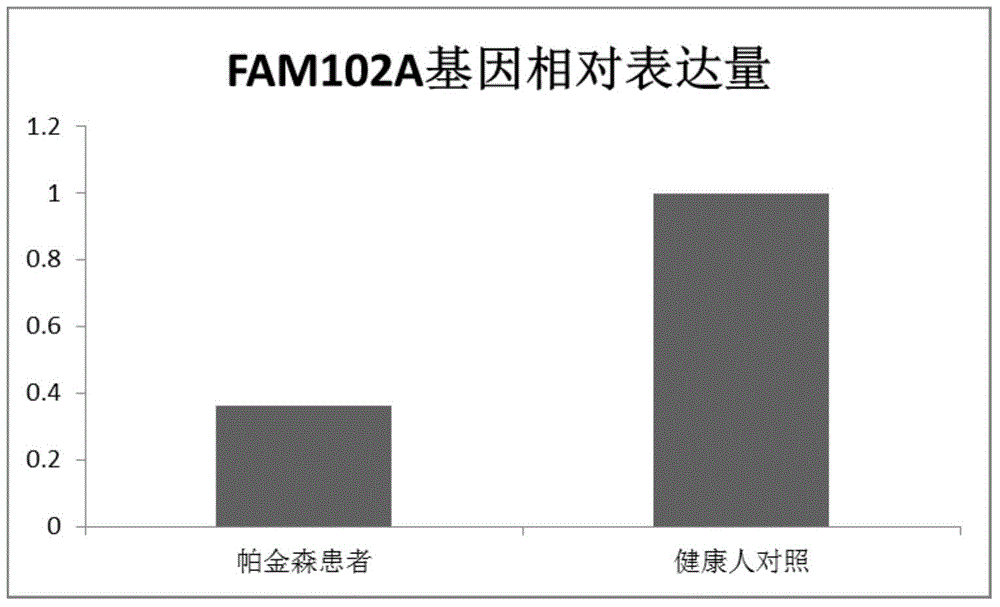
Parkinson diagnostic marker and application thereof
ActiveCN105063194AOrganic active ingredientsNervous disorderCandidate Gene Association StudyCandidate gene
The invention relates to a Parkinson diagnostic marker and application thereof, in particular to application of genes FAM102A to preparing Parkinson diagnosis and treatment reagents. The Parkinson diagnostic marker and the application have the advantages that genes are screened by an inventor on the basis of high-throughput sequencing results by the aid of bioinformatics methods and analysis, candidate genes FAM102A are selected, and relations between the genes FAM102A and Parkinson's diseases are proved by the aid of molecular biological methods; the genes FAM102A are in low-expression in the peripheral blood of Parkinson patients, have excellent correlation with Parkinson's diseases, can be used for preparing the auxiliary diagnosis and treatment reagents for the Parkinson's diseases and have important clinical application value.
Owner:QINGDAO MEDINTELL BIOMEDICAL CO LTD
Screening method and application of Dongxiang blue-eggshell chicken genome SNP molecular marker
PendingCN109295238AComprehensive and effective germplasm analysisPromote development and utilizationMicrobiological testing/measurementEggshellScreening method
Relating to the field of genetics, the invention specifically provides a screening method and application of a Dongxiang blue-eggshell chicken genome SNP molecular marker. The screening method includes: conducting high-throughput sequencing detection, then calculating the genetic statistics, then analyzing the genetic difference between Dongxiang blue-eggshell chickens and other chicken breed groups, and finally screening out the Dongxiang blue-eggshell chicken SNP molecular marker. The screening method has the technical effects of efficient and rapid screening of Dongxiang blue-eggshell chicken molecular marker, the selected gene of the screened Dongxiang blue-eggshell chicken serves as the molecular marker for identifying the Dongxiang blue-eggshell chicken germplasm, has the characteristics of low cost, abundant data and high detection precision, alleviates the technical problems of high genetic differentiation research cost and low accuracy caused by lack of efficient molecular biological method and genetic marker in current Dongxiang blue-eggshell chicken genetic differentiation identification. And application of the Dongxiang blue-eggshell chicken genome SNP molecular markeralleviates the technical problem of lack of efficient molecular biological marker in the genetic differentiation research of Dongxiang blue-eggshell chicken.
Owner:JIANGSU INST OF POULTRY SCI
Molecular biology method for identifying purity of tobacco varieties
InactiveCN105734141ANot affectedPreventing and ensuring the safety of production seedsMicrobiological testing/measurementBiotechnologyWhole genome sequencing
The invention relates to a molecular biology method for identifying purity of tobacco varieties. The method comprises the following main steps: respectively extracting total genome DNA of a control group and a to-be-detected tobacco variety, performing whole genome sequencing of proper depth by adopting the latest sequencing technology, performing sequence splicing, assembling and whole genome sequence comparison on the control group and the to-be-detected tobacco variety based on a tobacco genome reference sequence by utilizing bioinformatics means, counting base differences of the two groups, and calculating the purity percentage of the to-be-detected tobacco variety relative to the control tobacco variety. The method disclosed by the invention is not influenced by environmental conditions and seasons, is accurate and reliable in result, can accurately identify the purity of the tobacco varieties from a single base variation level of the smallest hereditary unit, can be used for parent purification and authenticity identification of the tobacco varieties and has great significances for guaranteeing safety of tobacco production varieties, maintaining economic benefits of tobacco growers and effectively solving intellectual property disputes of the tobacco varieties.
Owner:HUBEI TOBACCO SCI RES INST
Method for conversing switchgrass conducted by agrobacterium
InactiveCN101121940AImprove conversion efficiencyEfficient genetic transformation systemOther foreign material introduction processesPlant tissue cultureGreenhouseTransformation efficiency
The invention discloses an agrobacterium-mediated versatile-switch-grass transformation method. During the transformation, a man-induced young spike is taken as an explant; embryonic callus is induced on screening SMB culture medium; agrobacterium transformation and co-culture are adopted, and screening is conducted on culture media SM1 and SM2 successively to achieve resistant callus; the resistant callus is transferred to split-up culture media SMK1 and SMK2 successively to achieve regenerated plants; the regenerated plants are arranged in the rooting culture medium SMO and cultivated into seedlings; the seedlings are transplanted into a greenhouse or in a crop field for molecular biology-based testing; and the transgene plants expressed by extraneous objective gene is achieved according to molecular biologic method and objective character screening. The versatile-switch-grass transformation method has transformation efficiency about 10 percent, and the method lays a foundation for genetic improvement of the bio-energy (versatile switch grass) by transgene.
Owner:NORTHWEST A & F UNIV
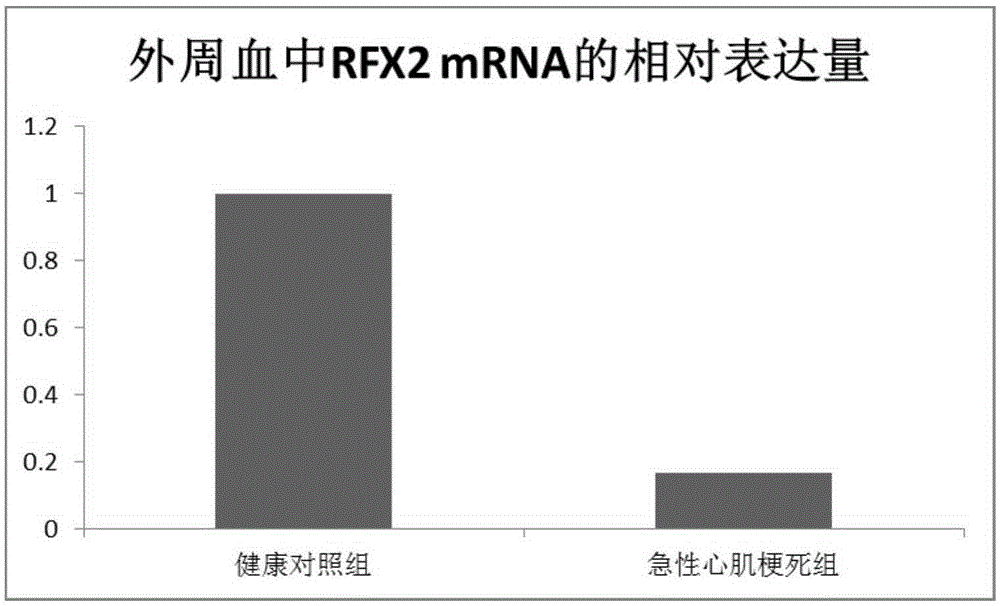
Acute myocardial infarction marker and application thereof
The invention relates to an acute myocardial infarction marker and an application thereof, and in particular relates to RFX2 and an application of an agonist thereof in preparing a medicine for diagnosis and treatment on acute myocardial infarction. Based on a high-flux sequencing result, the inventor screens out the RFX2 gene; a molecular biology method verifies that RFX2 has low expression in the peripheral blood and the cardiac muscle tissue of the acute myocardial infarction. The invention provides the potential diagnosis and treatment target spot of the acute myocardial infarction, and has great clinical application values.
Owner:QINGDAO MEDINTELL BIOMEDICAL CO LTD
A use of PD-0332991 in preparation of drugs preventing and treating drug-resistant tumor
ActiveCN104758292AGrowth inhibitionPrevent proliferationOrganic active ingredientsAntineoplastic agentsNude mouseGefitinib resistance
A use of PD-0332991 in preparation of drugs preventing and treating drug-resistant tumor is disclosed. Application of a cell biological method and a molecular biological method and human gefitinib-resistant nude-mouse transplanted tumor model tests prove that: the PD-0332991 can effectively inhibit growth of human gefitinib-resistant lung cancer cells, induce apoptosis, regulate and control cell cycle related gene, cause tumor cell cycle arrest and induce apoptosis. Accordingly, the PD-0332991 can be adopted as an effective component for preparation of drugs preventing and treating gefitinib-resistant tumor, and foods, health products and cosmetics for preventing and treating drug-resistant tumor.
Owner:GENERAL HOSPITAL OF TIANJIN MEDICAL UNIV
Molecule identification and transfer technology for broad-spectrum rice-blast resistant gene of paddy rice
InactiveCN1840696AImprove use valueImprove breeding efficiencyMicrobiological testing/measurementDiseaseResistant genes
The related rice gene mark for broad spectrum anti rice blast includes mark length 10-30 nucleotides with sequence same as the sequence of 1st-70798th in SEQ ID No:1. Wherein, based on our rice resource and natural or induced diseases, it applies different rice blast plants with the domestic as main and foreign as assist to screen the 'Gumei 4th', uses molecular mark technique to locate gene and obtain Pi-gm(t) gene with chain mark, and has great application value in breeding.
Owner:CHINA NAT RICE RES INST +1
Molecular biology method for quickly identifying Heliothis armigera and Helicoverpa assulta
The invention discloses a method for quickly identifying Heliothis armigera and Helicoverpa assulta, and belongs to the field of molecular biology. The method comprises the following steps of: extracting total DNA by adopting a standard phenol-chloroform protocol or a DNA extraction kit; amplifying a front sequence of mitochondria CO1 by using a DNA barcode primer, and using the amplified sequence as a mark; and performing diagnostic characters of nucleotides. By the method, different biotypes and larvae or incomplete individuals of the same variety of Heliothis armigera and Helicoverpa assulta can be identified; and the method has the characteristics of quickness and convenience, and is more accurate and reliable than the traditional morphologic identification.
Owner:HONGYUN HONGHE TOBACCO (GRP) CO LTD
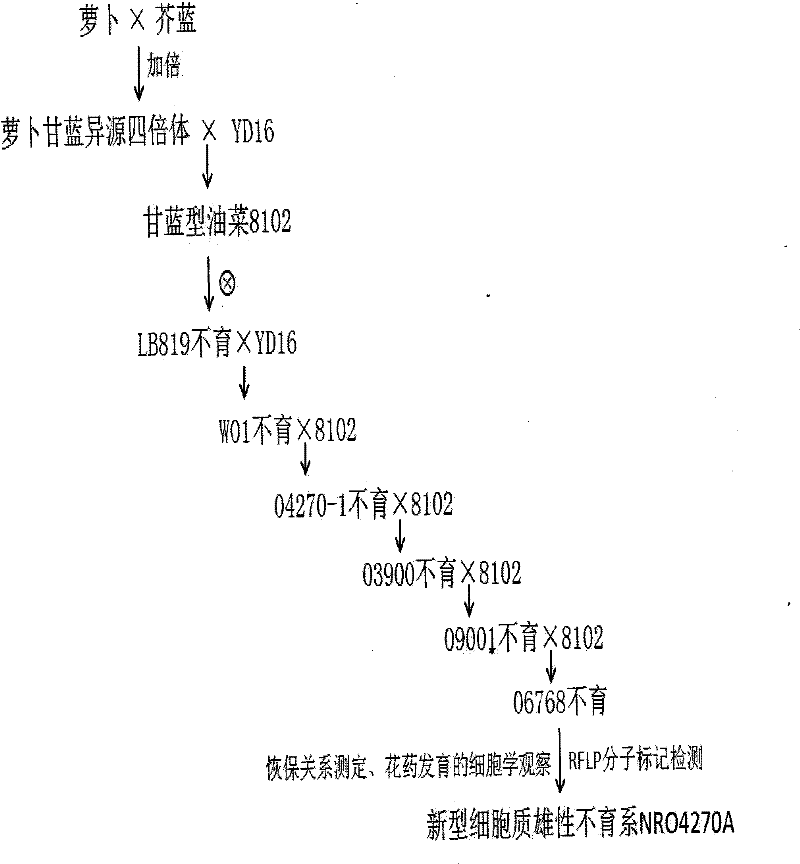
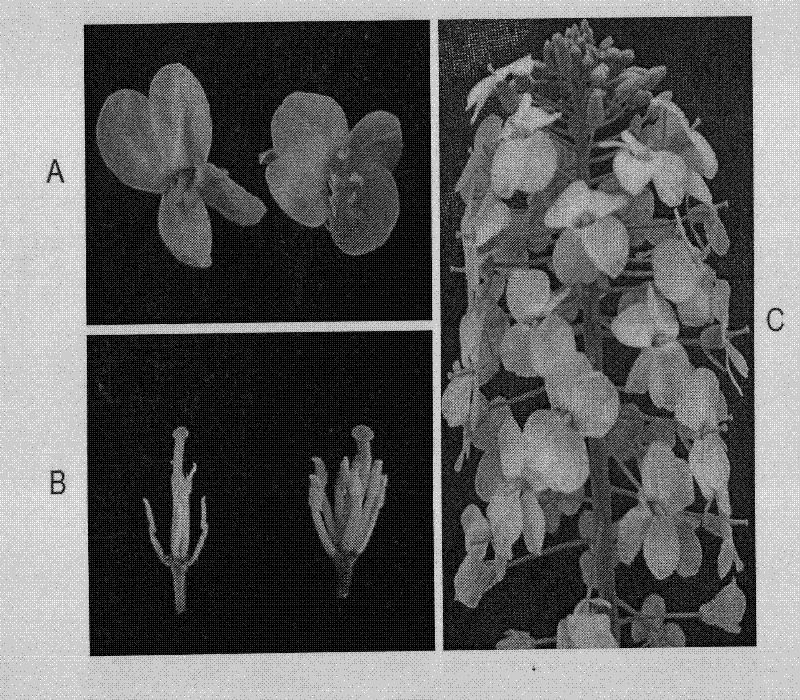
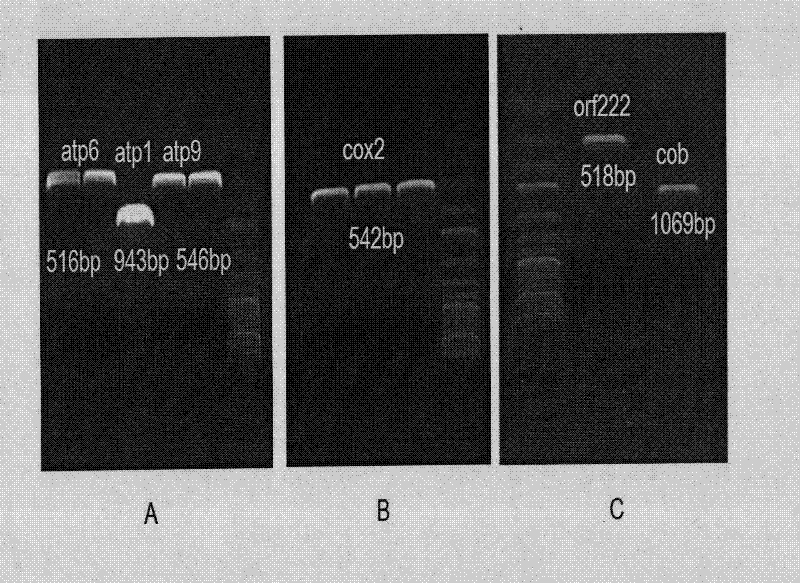
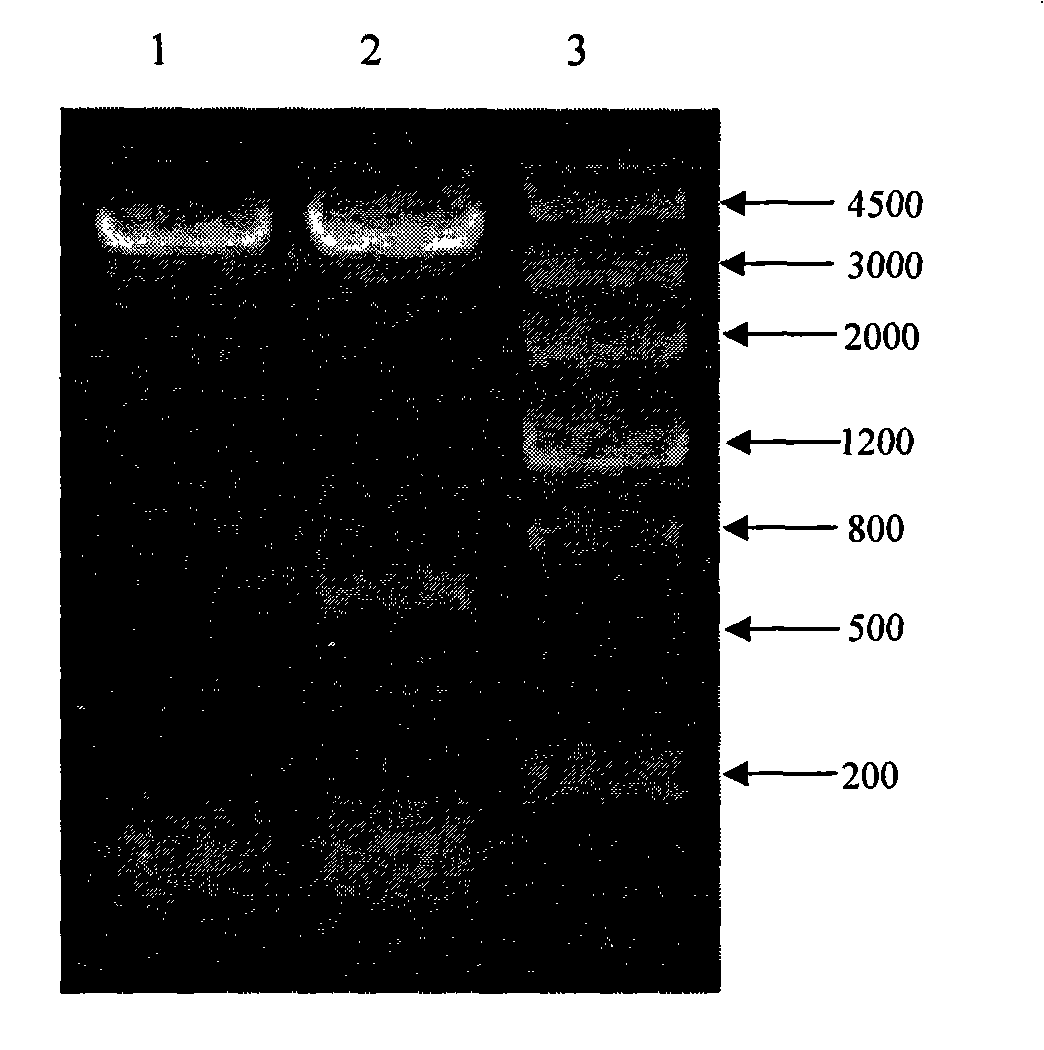
Selection breeding method for a Brassica napus-turnip cabbage cytoplasmic male sterility line
InactiveCN102475057AGenetically stable sterilityExcellent agronomic traitsPlant genotype modificationSelfingCytoplasmic male sterility
The invention belongs to the technical field of rape breeding, and discloses a selection breeding method for a Brassica napus-turnip cabbage cytoplasmic male sterility line (CMS). The method is characterized in that an artificially-bred turnip cabbage allotetraploid as a female parent is hybridized with a Brassica napus variety as a male parent, male sterile plants are isolated from the progeny of hybrid selfing, and a Brassica napus-turnip cabbage cytoplasmic male sterility line with genetic stability is obtained by successive backcross breeding. Restorer line and maintainer line relationship and molecular biological methods are employed to prove genetically and on DNA level respectively that the Brassica napus-turnip cabbage cytoplasmic male sterility line has a sterile cytoplasmic type different from those of Polima cytoplasmic male sterility (pol CMS), turnip cytoplasmic male sterility (ogu CMS, kos CMS) and Brassica juncea cytoplasmic male sterility (hau CMS). The invention adds a new rape cytoplasmic male sterility type for China and the rest of the world.
Owner:HUAZHONG AGRI UNIV
Recombined chicken alpha interferon gene and recombinant vector thereof
InactiveCN101338314AStrong antiviral activityEase of mass productionFungiMicroorganism based processesHigh concentrationAnti virus
The invention relates to a novel gene sequence of a recombinant chicken Alpha interferon, constructs a recombinant expression vector thereof and belongs to a gene engineering biological product obtained by a molecular biology method. The gene sequence of a newly designed chicken Alpha interferon is recombined into a pPICZ Alpha-A vector and then is confirmed on the special position of a microzyme by an electric conversion mode. Besides, a pichia expression system is adopted to express a foreign gene, thus being beneficial to the commercial production of the chicken interferon. The protein expressed by a gene group after being diluted by 4365158.3 times can completely restrain the attraction of vesicular stomatitis virus of 100-1000TCID50. Compared with a natural chicken Alpha interferon, the novel gene sequence of a recombinant chicken Alpha interferon has a higher anti-virus effect; the anti-virus effect thereof is improved by 8 times. Test also detects that the protein expressed by the gene can restrain the proliferation of a newcastle disease virus and an avian influenza virus; besides, the effect of the interferon with high concentration is more remarkable.
Owner:NANJING AGRICULTURAL UNIVERSITY
Efficient genetic transformation method of hybridized tulip tree
ActiveCN103088059AReduce generationEasy to importFermentationGenetic engineeringSomatic embryogenesisTulip Trees
The invention discloses an efficient genetic transformation method of a hybridized tulip tree. The efficient genetic transformation method comprises the following steps of: setting up a hybridized tulip tree transformation receptor, preparing agrobacterium, infecting, culturing without bacterium and culturing in a screening manner. According to the efficient genetic transformation method, unicell subjected to the suspension cultivation of the hybridized tulip tree serves as a genetic transformation acceptor system; the GUS gene is regarded as a report gene through a southern buddhism; and transient expression and long-term expression of the GUS gene are determined through a cytological method and efficient transformation of foreign gene is determined by detecting through a molecular biological method. As a somatic embryogenesis system generated by the unicell of the hybridized tulip tree is adopted, the efficient genetic transformation method of the hybridized tulip tree is good for leading in the foreign gene, and reduces generation of chimera.
Owner:NANJING FORESTRY UNIV
Performance improved recombination staphylococcus aureus protein A affinity ligand and construction method thereof
ActiveCN103214563AImprove bindingImprove elutionMicroorganism based processesDepsipeptidesHigh concentrationEscherichia coli
The present invention discloses a performance improved recombination staphylococcus aureus protein A affinity ligand and a construction method thereof. The present invention adopts a molecular biology method. A sequence B of a nature protein A is selected for molecular transformation. A C-terminal of the sequence B is added with two cysteines, so that the protein A can pass through double-locus coupled chromatography matrix to stabilize the connection. Six glycines are added to the end of a second Loop of the sequence B to increase the length and reduce the binding force with an antibody, so that elution conditions are mild. On this basis, resistance performance to high concentration base of the protein A is transformed. Asparagines and phenylalanine at 23rd and 30th positions of the sequence B are respectively replaced by threonine and alanine to obtain a sequence Z with higher alkaline resistance properties. Then, isocaudarner is used for connecting sequence Zs of different numbers head-to-tail in series. The efficient expression system of e. coli is used for overexpression. The expressed recombination protein A is coupled to agarose matrix preparation affinity chromatography fillers and is used for purifying antibodies. Results show that the recombination protein A affinity ligand prepared by the present invention is good in elution performance and alkali resistance.
Owner:嘉兴千纯生物科技有限公司
Phospholipase B from pseudomonas fluorescens and production method thereof
InactiveCN102002486AHigh activityImprove stabilityHydrolasesMicroorganism based processesEscherichia coliEnzyme Gene
The invention relates to a phospholipase B from pseudomonas fluorescens and a production method thereof, which belong to the field of enzyme gene engineering and enzyme engineering. The phospholipase B is produced from the pseudomonas fluorescens, has the total gene length of 1272bp and codes of 423 amino acids, and the zymoprotein theoretical molecular weight is 45.8kDa, wherein 1 to 69bp code phospholipase B signal peptide, and 70 to 1272bp code phospholipase B mature peptide are comprised. An expression vector and a recombinant host of the phospholipase B can be obtained by the traditional molecular biological method, i.e. colibacillus recombinant plasmid and recombinant colibacillus containing genes of the phospholipase B are obtained. The phospholipase B has good activity and stability at low temperature, does not have the lipase activity, can be hydrolyzed for catalyzing two fatty acyl group radicals in complete phospholipids, has wide application in the industrial process of grease refining, phospholipids emulsifying agent modification and the like, and also has simple preparation method, high product quality and low production cost.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY
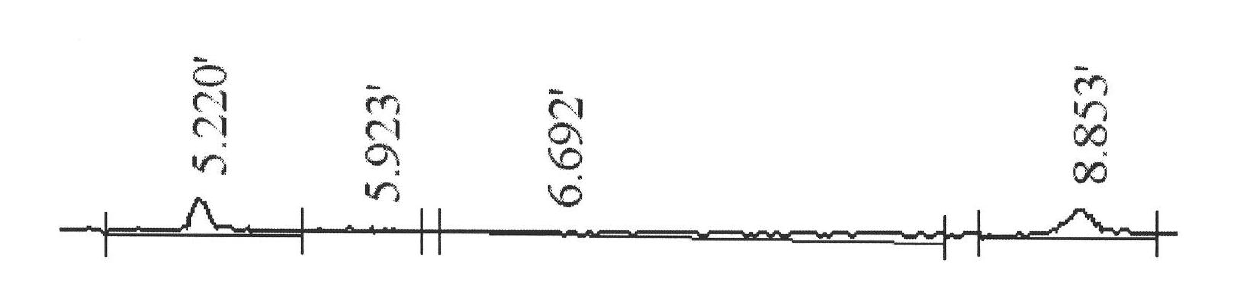
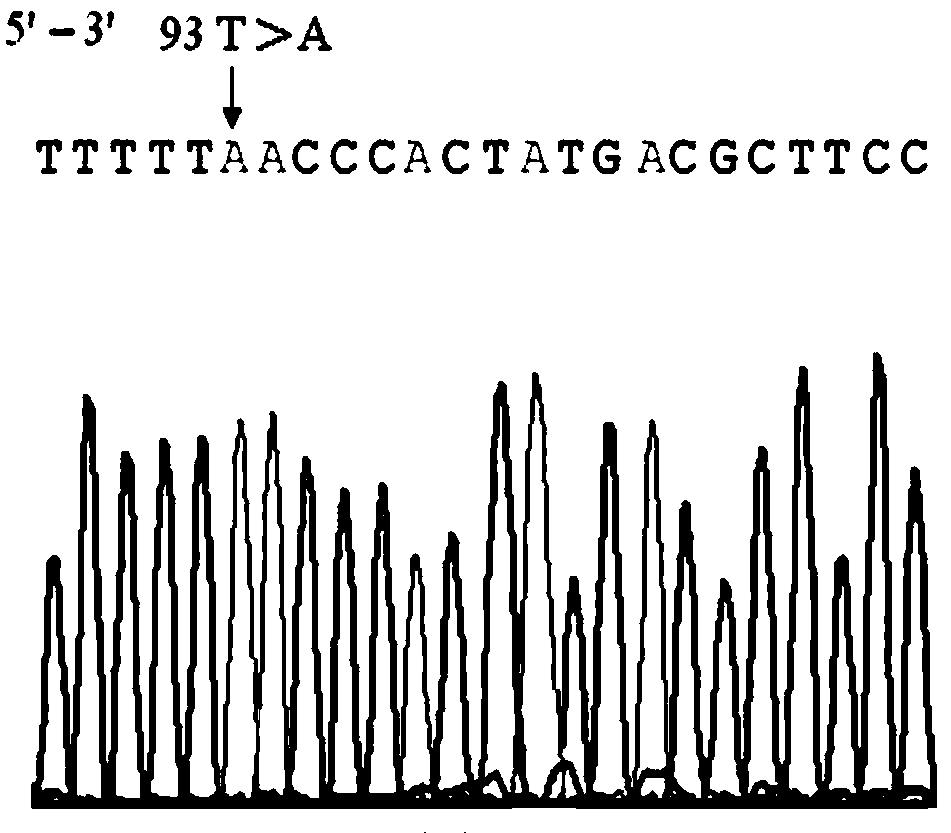
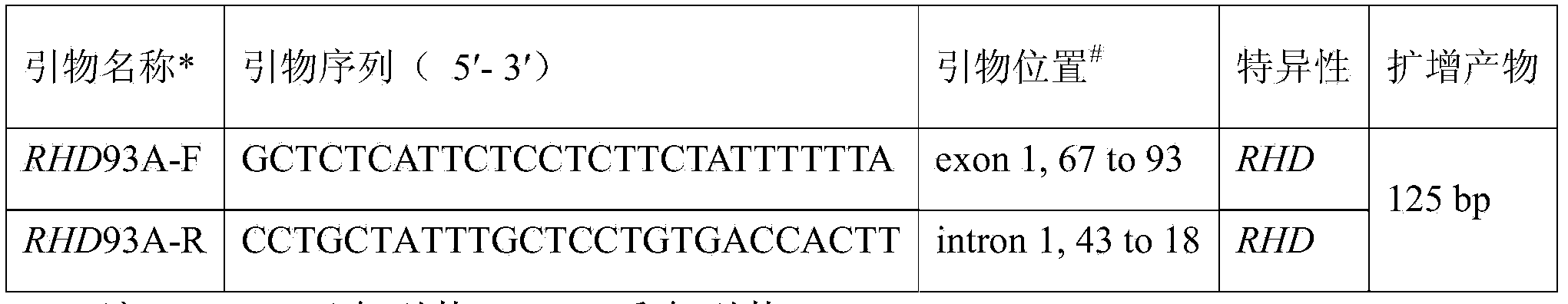
RH blood type DEL-type RHD93T>A allele and detection method thereof
InactiveCN103966228AExtensive scientific research application valueMicrobiological testing/measurementFermentationRegioselectivityRhD antigen
The invention relates to an RH blood type DEL-type RHD93T>A allele and a detection method thereof, and belongs to the technical field of molecular biology analysis and detection. According to the detection method, primer sequences capable of amplifying areas including target sites with mutant gene sequences are designed through a PCR-specific primer technology; PCR amplification is selectively conducted on the areas of the target sites including the mutant genes through a gene amplification method so as to specially detect specific RHD93T>A allele. The detection method adopts a molecular biology method for gene-level detection, so that the detection sensitivity and specificity are improved, and the detection method is simple, convenient and fast. Due to differences in RHD gene of different ethnic groups, the detection method is designed specially for RHD93T>A allele on the basis of correlational research and according to the Chinese RHD gene molecule background. The RH blood type DEL-type RHD93T>A allele and the detection method thereof not only are of important clinical practical significance in making up for deficiencies of the serological technique, but also has a wide application value on scientific research.
Owner:WUXI NO 5 PEOPLES HOSPITAL
Molecular-biological method for quickly distinguishing noctuidae pests
InactiveCN102373292AShorten the timeLow costMicrobiological testing/measurementBiotechnologyNucleotide
The invention discloses a molecular-biological method for quickly distinguishing prodenia litura and other two noctuidae pests (cotton bollworm and tobacco budworm). The method comprises the following steps of: extracting total DNA (Deoxyribonucleic Acid) by adopting a standard phenol-chloroform method or a DNA extraction kit; using a DNA bar code primer to amplify a sequence at the front part of mitochondria CO1 as a label; and comparing diagnostic characters of nucleotides. The molecular-biological method can be used for distinguishing different biotypes and larvae or incomplete individuals of the prodenia litura, the cotton bollworm and the tobacco budworm, has the characteristics of accuracy, quickness, convenience and quickness, and is more accurate and reliable compared with the traditional morphological identification.
Owner:YUNNAN NORMAL UNIV
Promotor replacement method for improving volume of production of bacillus subtilis surfactin
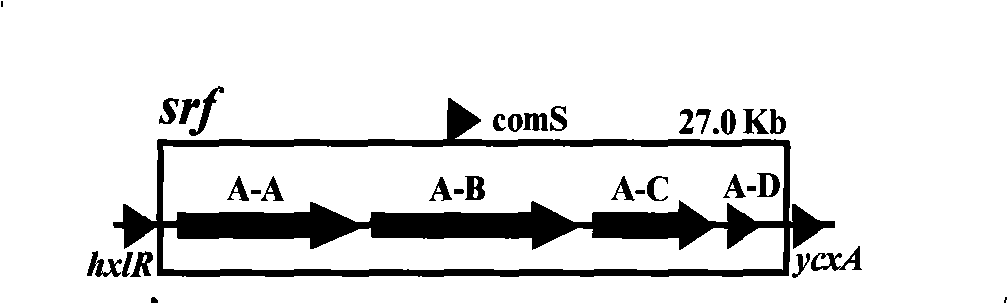
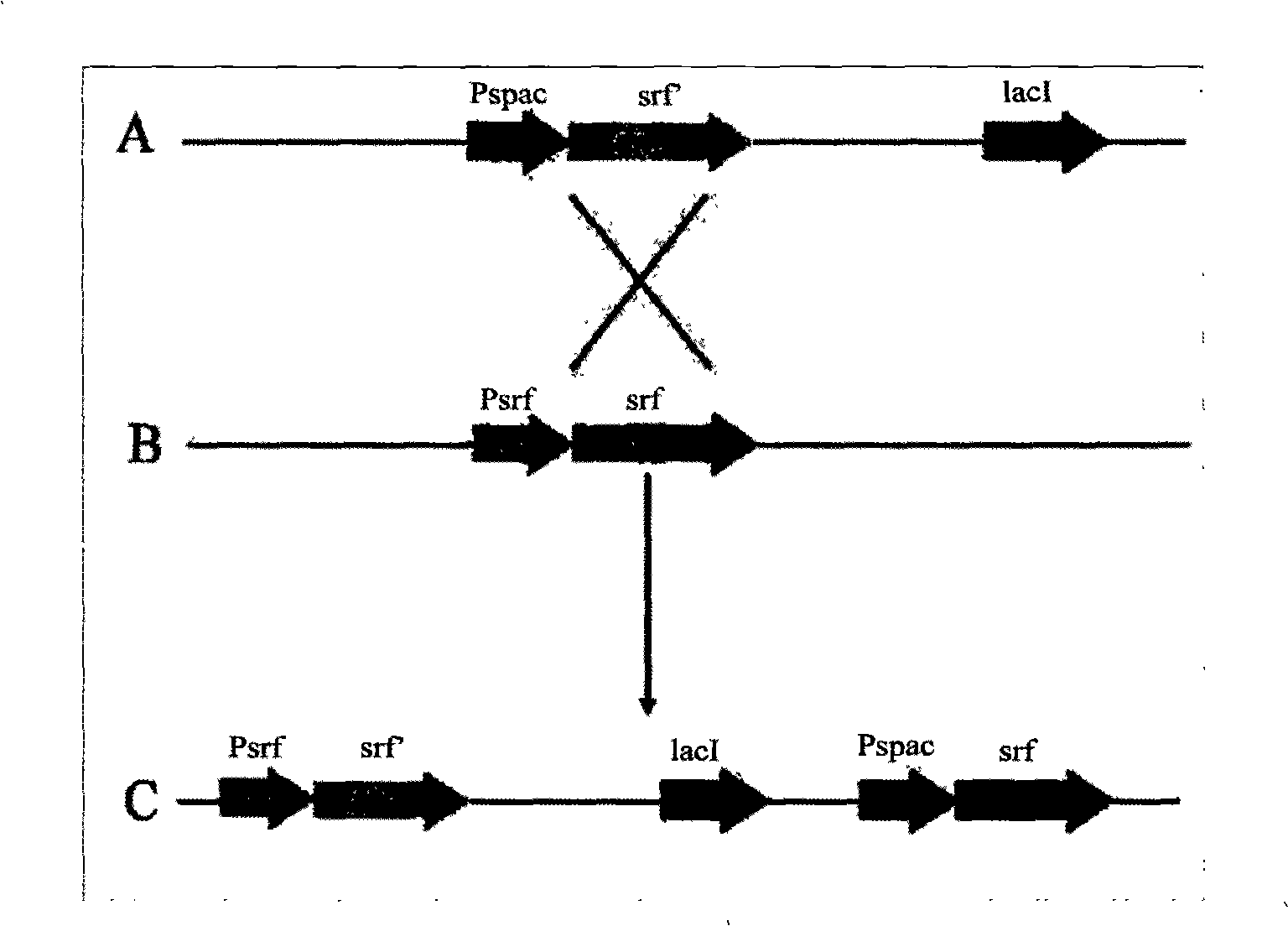
InactiveCN101402959AIncrease productionEasy to produceBacteriaMicrobiological testing/measurementBiotechnologyAntimikrobielle peptide
The invention relates to a promoter replacement method for improving the yield of Bacillus subtilis surfactin, belonging to the biotechnology field. The DNA homologous integration technology is adopted for allowing a promoter Pspac to replace the promoter of a Bacillus subtilis fmbR surfactin synthase gene. The homologous sequence of 1081bp is obtained by amplification from an fmbR strain genome through the PCR method and connected to HindIII and BamH I restriction enzyme cutting sites of a plasmid pMUTIN4, a well constructed vector is converted into wild Bacillus subtilis fmbR, thereby realizing the replacement through the screening by an antibiotic culture medium and the molecular biology method. The method can directly improve an antimicrobial peptide production strain on genetics, thereby having potential application value in industry. The yield of the strain surfactin is improved by about 4.85 times, and the yield of the surfactin can be improved by about 10 times under the IPTG induction.
Owner:NANJING AGRICULTURAL UNIVERSITY
Culicoides bellulus specific gene and molecular identification method thereof
InactiveCN105543249AGuaranteed accuracyEnsure reliabilityMicrobiological testing/measurementEnzymesMolecular identificationCulicoides
The invention discloses a culicoides bellulus specific gene and a molecular identification method thereof. The culicoides bellulus specific gene is characterized in that a molecular biology method is adopted to obtain the COI gene sequence of culicoides, and the species of the culicoides is identified by comparing gene sequence similarity. The ulicoides bellulus molecular identification method is beneficial for accurately and fast identifying culicoides bellulus.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
Primers, kit and detection method for detecting gene type of dominant white feather site of chicken PMEL17 gene
ActiveCN103602745AAccurate detectionQuick checkMicrobiological testing/measurementDNA/RNA fragmentationDominant whiteGene type
Owner:HENAN AGRICULTURAL UNIVERSITY
Human papillomavirus type bivalent virus-like particle mixed protein antigen and construction method
InactiveCN1683010AImproving immunogenicityImprove responseAntiviralsRecombinant DNA-technologyVaccine ImmunogenicityIndividual animal
The present invention relates to human papillomavirus type bivalent virus-like particle mixture, and human papillomavirus type bivalent virus-like particle mixing protein antigen and its construction method. The present invention includes one kind of cervical carcinoma virus HPV16 L1 antigen and one kind of condyloma acuminata virus HPV11 L1 antigen. The HPV11 L1 gene is first cloned from condyloma acuminata tissue through molecular geological method, and both HPV16 L1 gene cloned from cervical carcinoma tissue and the HPV11 L1 gene are then expressed in the insect cell expression system of baculovirus, so as to constitute recombinant virus strain Bacmid / HPV16-HPV11L1. Animal immunizing test shows that the constituted recombinant protein has excellent immunogenicity and immunoreactivity, and may be used as candidate antigen of gene engineering multivalent vaccine for preventing condyloma acuminata and cervical carcinoma.
Owner:HARBIN MEDICAL UNIVERSITY
Multiplex PCR method for rapid detection and identification of five Listeria species
InactiveCN102286631AHigh sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesForward primerMultiplex
The invention discloses a method for multiplex polymerase chain reaction (PCR) fast detection and five-Listeria-bacterium identification, which comprises the following steps of: 1, firstly selecting five forward primers and one reverse primer; 2, taking positive control standard bacterium liquid and bacterium liquid to be detected for being respectively prepared into genome deoxyribonucleic acid (DNA) templates; 3, respectively adding the forward primers and the reverse primer into the genome DNA templates prepared by the positive control standard bacterium liquid and the genome DNA templates prepared by the bacterium liquid to be detected for carrying out PCR amplification reaction; 4, taking amplified products for carrying out electrophoresis; and 5, judging the results. The method provided by the invention belongs to the fast, accurate and sensitive method for the multiplex PCR detection and the five-Listeria-bacterium identification, and the selection is provided for the identification of the Listeria bacterium by a molecular biological method. The efficiency is improved, the time is saved, and in addition, the detection cost is saved.
Owner:INSPECTION & QUARANTINE TECH CENT OF XIAMEN ENTRY EXIT INSPECTION & QUARANTINE BUREAU
Probe and method for detecting red naked dinoflagellate
InactiveCN1699598AObjective detectionAccurate detectionMicrobiological testing/measurementMicroorganismNucleic Acid Probes
The present invention discloses the oligonucleotide probe based on the design of red gymnodinia ribosome DNA (rDNA) nucleic acid sequence, belonging to the technical field of environment microbe inspection by means of molecular biological method. This nucleic acid probe is used as a pair of guide objects to carry out PCR reaction for accurate and fast inspection of red gymnodinia through predicting whether there is PCR product of corresponding size. The present invention also discloses a fast and accurate inspection and quantitative counting method of red gymnodinia by fluorescent quantitative PCR technology with the probe.
Owner:OCEAN UNIV OF CHINA
Method for the design of oligonucleotides for molecular biology techniques
InactiveUS20070059743A1Microbiological testing/measurementBiological testingSequence databaseOrganism
The present invention discloses a method that can be used to identify one DNA sequence or one specific group of DNA sequences from a complex biological sample. Diverse molecular biology methods require the use of short DNA sequences, called oligonucleotides, that are artificially synthesized from a description of their composing bases. The disclosed method allows the design of oligonucleotides useful for said molecular biology procedures, like probe design procedures, and is characterized by the construction of a database of reference sequences, the selection of a subset of sequences belonging to target organisms, the selection of candidate oligonucleotides from such sequences, the depuration of these candidate oligonucleotides according to hybridization specificity and thermodynamic stability criteria, and the sorting of such oligonucleotides according to their taxonomic specificity. In a second aspect, a method is disclosed to design oligonucleotides pairs or primers, which are required in certain molecular biology techniques, like polymerase chain reaction (PCR) techniques. This method is similar to the first aspect of the invention, but thermodynamically compatible oligonucleotides pairs or primers that hybridize to the same sequence at a distance which is within a given range are evaluated.
Owner:BIOSIGMA
Application of isothiocyanate to preparation of medicine for preventing and treating medicine-resistance tumor
ActiveCN101822663AGrowth inhibitionEster active ingredientsAntineoplastic agentsBULK ACTIVE INGREDIENTWilms' tumor
The invention discloses application of isothiocyanate to the preparation of medicine for preventing and treating medicine-resistance tumor. The invention uses a celluar and molecular biology method for proving that the isothiocyanate can effectively inhabit the growth of medicine-resistance tumor cells, induce the apoptosis, regulate and control the tumor medicine-resistance relevant genes, cause medicine-resistance tumor cell cycle arrest, and cause tumor cell oxidative damage. Thereby, the isothiocyanate can be used as a active ingredient for preparing the medicine for preventing and treating medicine-resistance tumor, and food, health-care products and cosmetics for preventing and treating medicine-resistance tumor.
Owner:GENERAL HOSPITAL OF TIANJIN MEDICAL UNIV
Reagent composition for separating total RNA in plant or microorganism and preparation method thereof
InactiveCN102443580AQuality improvementOvercoming selectivityDNA preparationSodium acetatePrecipitation
The invention belongs to the field of molecular biology method and relates to a reagent composition for separating total RNA in a plant or a microorganism and a preparation method thereof. The high quality RNA can be obtained after a simple extraction by chloroform while guanidinium isothiocyanate and phenol are regarded as main components, positive ions are provided by NaCl, MgCl2 and the like, and a solution pH is stabilized by a sodium acetate-acetic acid buffer system. Glycogen serving as a nucleic acid precipitant is added in the RNA extraction reagent in the invention, so that the reagent disclosed by the invention, in comparison with reagents of the same type, greatly improves precipitation efficiency of the nucleic acid and also can efficiently separate the nucleic acid component which has a very low content in a tissue sample. Therefore, the nucleic acid precipitation process can be finished in a short period, the precipitation process at -20 DEG C for hours is avoided, and operating time is shortened greatly. The method is rapid as well as efficient and has wide applicable samples; and the reagent composition disclosed by the invention is cheaper than commercial TRIzol reagents. The disadvantages of high sample selectivity, fussy operation and relatively low efficiency of the traditional method are overcome. The operation is more flexible; and the sample after being homogenized can be stored at -20 DEG C and then used for the RNA extraction.
Owner:HUAZHONG AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com