Method for obtaining SSR (Simple Sequence Repeat) primer of lycium ruthenicum based on transcription sequencing
A technology of Lycium nigra and transcriptome sequencing, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of the lack of genome information and the small number of SSR primers, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
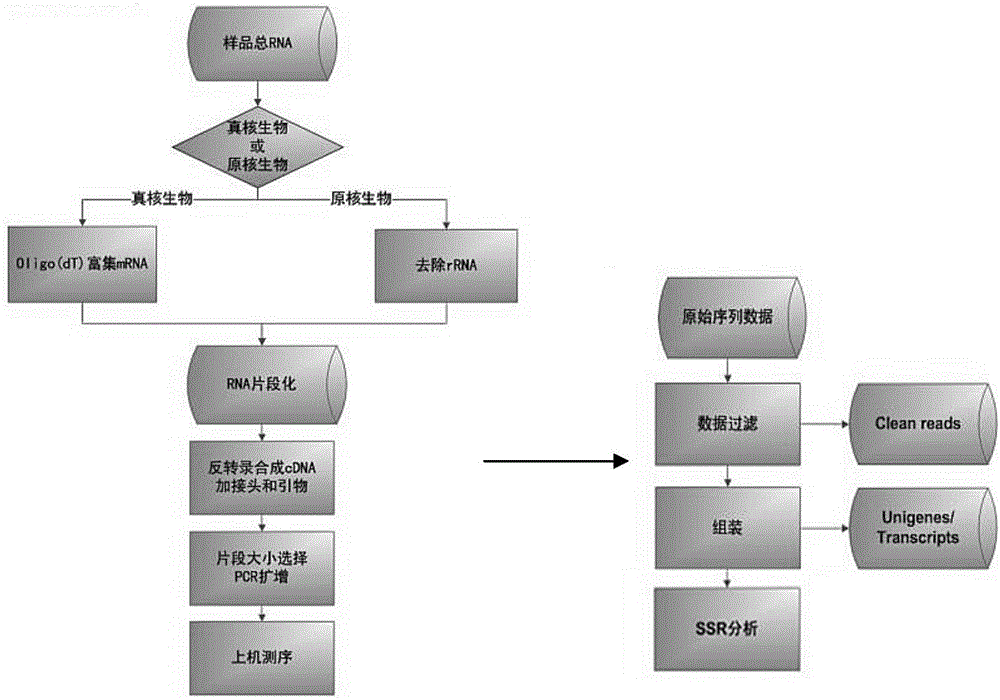
[0078] A method for obtaining SSR primers of Lycium barbarum lycium barbarum based on transcriptional sequencing, the steps are as follows:
[0079] 1) Take the black fruit Lycium barbarum from 3 provenances as samples, and perform transcriptome sequencing on the above-ground parts, and divide them into control group, salt treatment group and mixed salt treatment group. The salt treatment group uses 100mL, pH=7, 200mmol / L NaCl solution, mixed salt treatment group used 100mL, pH=8.54, NaCl 2 CO 3 , NaCl, Na 2 SO 4 , NaHCO 3 According to the mixed solution with a final concentration of 200mmol / L mixed according to 1:9:9:1, after 6 hours of treatment, the leaves were frozen and stored in liquid nitrogen for transcriptome sequencing;
[0080] 2) Extract the total RNA of Lycium barbarum by CTAB method, separate the mRNA, combine it into double-stranded cDNA, add adenine and sequence linker, amplify the fragments with a size of 200bp-700bp, and use Illumina HiSeq 2000 to analyz...
Embodiment 2
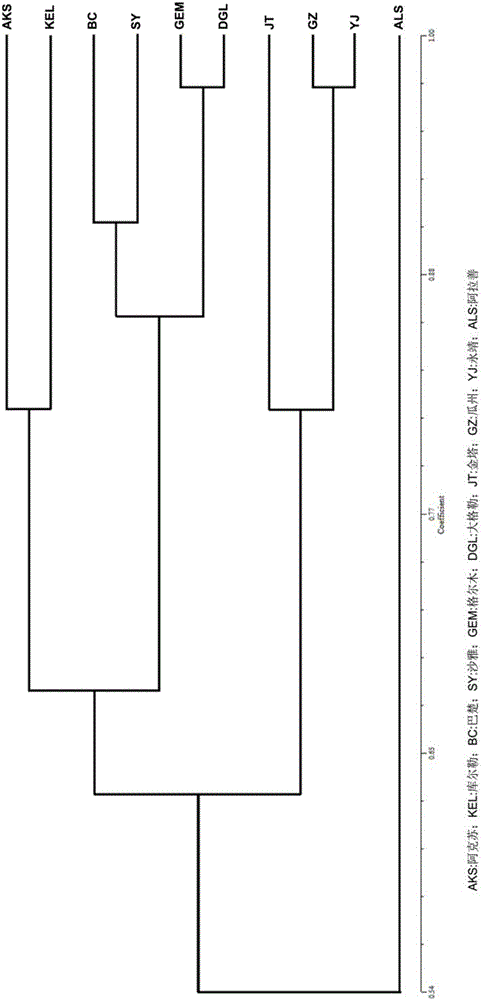
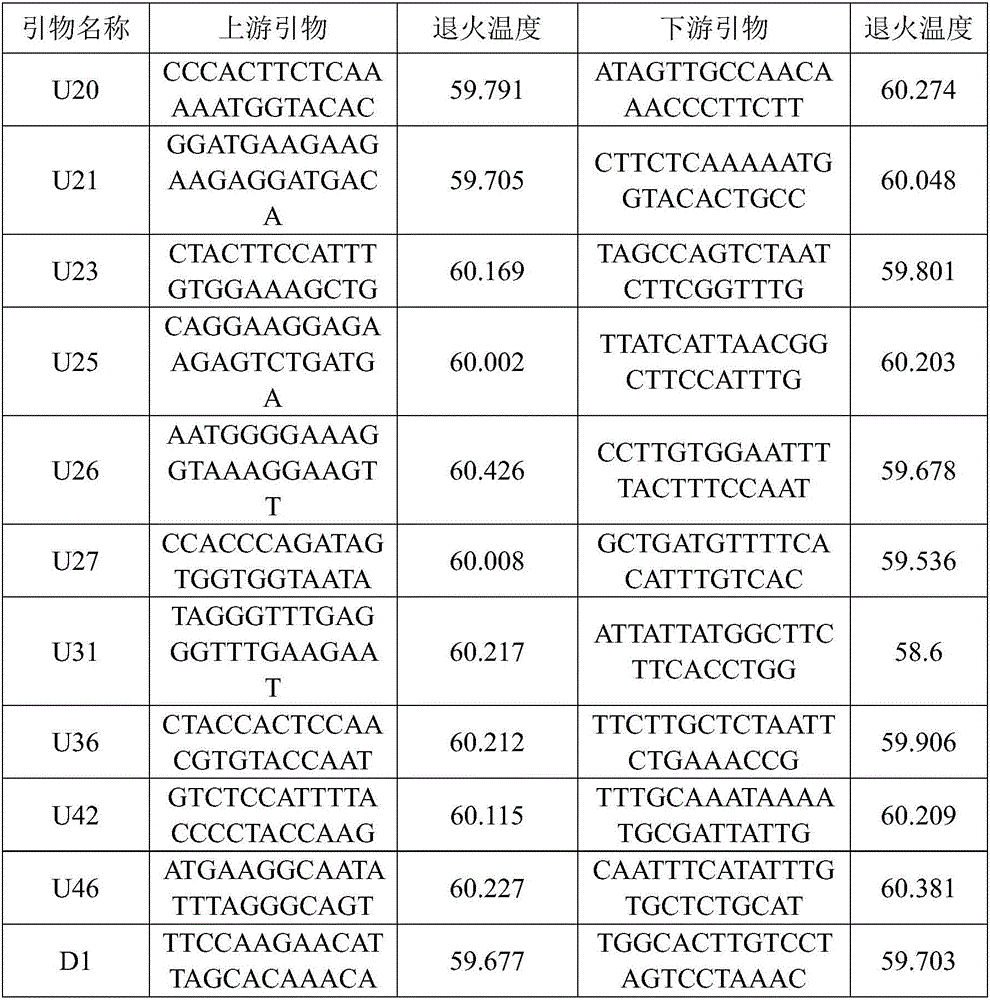
[0100] Example 2 Utilize SSR primers to identify Lycium barbarum
[0101] 1. DNA extraction
[0102] Using the CTAB method to extract the DNA of 10 provenances of Lycium barbarum black fruit, 25 samples were taken from each provenance as repetitions, the specific steps are as follows:
[0103] ①Take the leaf sample stored at ultra-low temperature, grind it in a mortar and add it to a 2.0ml centrifuge tube (the amount of leaf powder added should not exceed the tip of the bottom of the 2.0ml centrifuge tube;
[0104] ②Quickly add 500 μl of CTAB preheated at 65°C (2% CTAB, add 0.5-1% β-mercaptoethanol before use), mix well, put it in a water bath at 65°C for half an hour, and shake it every ten minutes to make CTAB and leaves Mix the sample thoroughly;
[0105] ③Take out the centrifuge tube and let it stand for 2 minutes, add an equal volume of chloroform / isoamyl alcohol (24:1) mixture, mix the mixture with CTAB, and vortex to mix, so that CTAB and chloroform can fully contact;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com