Molecular marker of pot shattering resistance trait major gene locus of rapes and application
A molecular marker and major gene technology, applied in the fields of molecular biology and genetics and breeding, can solve the problems that quantitative traits are easily affected by environmental conditions, long cycle, poor selection effect, etc., and achieve a convenient and fast detection method with high cost. , the effect of improving the selection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1. Construction of Rapeseed Anti-crack Angle Segregation Population and Determination of Characters
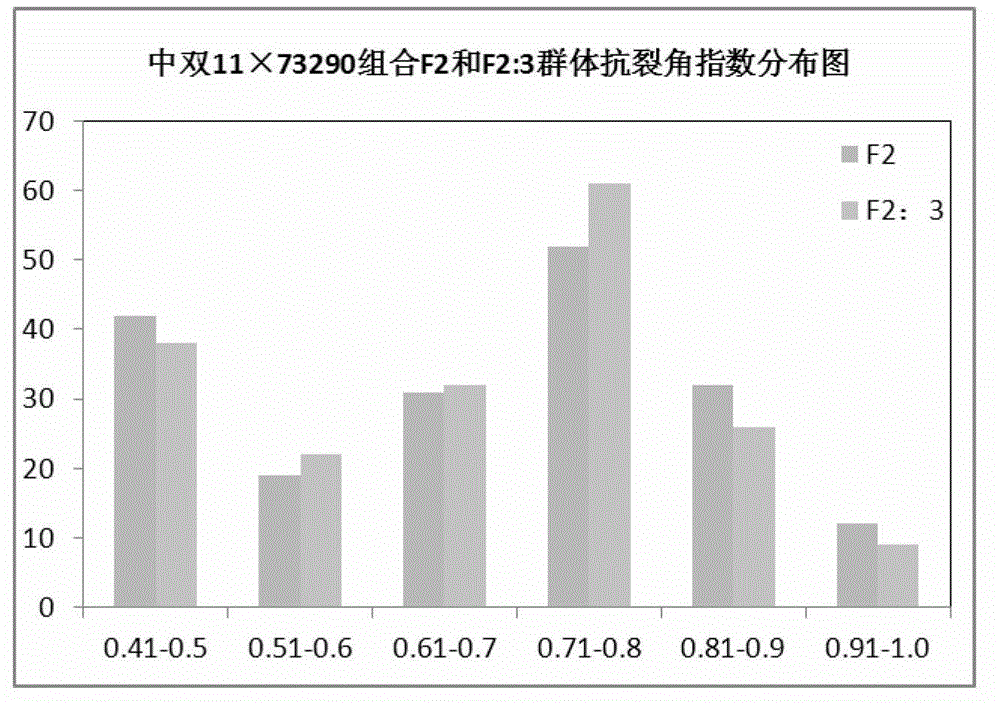
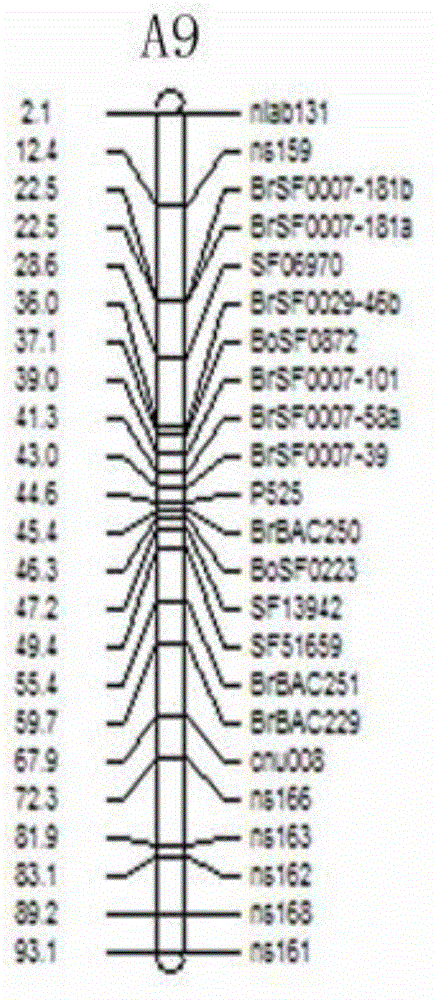
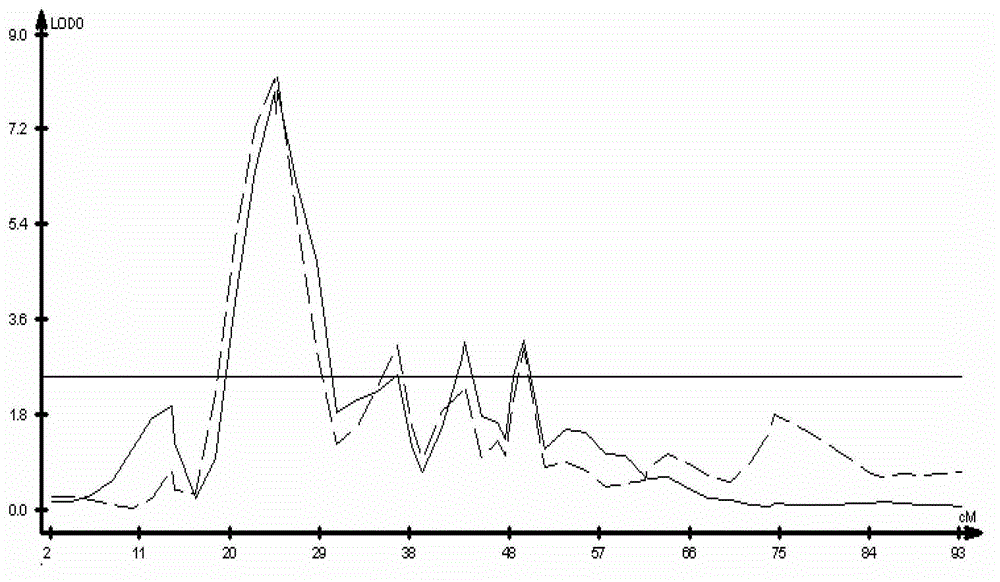
[0032] The populations used in this example are F2 and F2:3 populations of the offspring of rapeseed with high and no resistance to cleavage angle (Zhongshuang 11: cleavage angle resistance index 0.83; 73290: cleavage angle resistance index 0.48). The cleavage-horn resistance phenotypes of the parents, F2 and F2:3 populations were identified after harvesting at maturity. The results of anti-cracking angle index of F2 and F2:3 segregation populations showed that: the anti-cracking angle index of the two populations showed a continuous distribution, but the variation distribution did not show a typical normal distribution, which proved that the anti-cracking angle trait was a quantitative trait and there was a main effect gene locus ( figure 1 ).
Embodiment 2
[0033] Example 2. Parents, F2 and F2:3 Extraction of Total DNA of Segregation Population Leaves
[0034] The total DNA of leaves was extracted by the CTAB method, and the specific steps were as follows:
[0035] A. Take 0.1 gram of rapeseed leaf fresh sample and grind it, add 700 microliters of extract to grind it, then put it into a 1.5 milliliter centrifuge tube and place it in a constant temperature water bath at 65°C for 60 minutes, during which it mixes 2-3 times;
[0036] B. Add an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1, V / V / V), invert gently to mix well; centrifuge at 12000rpm for 10 minutes to make layers, and gently suck the supernatant into another 1.5 ml centrifuge tube; add an equal volume of chloroform: isoamyl alcohol (24:1, V / V) and re-extract once;
[0037] C. Add 1 ml of -20°C pre-cooled absolute ethanol, freeze at -20°C for no more than 30 minutes to allow DNA precipitation; centrifuge at 12,000 rpm for 10 minutes, discard the ethano...
Embodiment 3
[0039] Example 3. Development and synthesis of primers and screening of polymorphisms
[0040] The SSR primers we used include two types: one is the primer sequences published in published articles and Brassica database (http: / / www.brassica.info / resource / markers / ssr-exchange.php), including CB, CNU, niab, MR, FITO, Ol, BRMS, BRAS, Ni, Na, BN, NGA, MB, BnGMS, BrGMS, BoGMS, BnEMS, and many other series; the other type is spliced by our sequencing of cabbage and cabbage The scaffold sequences were developed and named BrSF and BoSF series respectively. The specific development method is to use SSRHunter software to search for SSRs in each scaffold, and then use Primer3.0 software to design SSR primers. We have synthesized a total of 3085 pairs of public and newly developed SSR primers through the biological company, and the screening results showed that 15% of the amplified products of the primers had length polymorphism differences between the parents. The polymorphism scree...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com